Abstract
N6-methyladenosine (m6A) RNA methylation has recently been found involving in regulatory mechanism of the tumor progression. Our aim was to explore the biological function and clinical significance of the m6A methyltransferase METTL3 in intrahepatic cholangiocarcinoma (ICC). In this study, we revealed that METTL3 was upregulated and predicted poor prognosis of patients with ICC. Multivariate regression analysis demonstrated that METTL3 expression was an independent predictor for overall survival in patients with ICC. Moreover, METTL3 knockdown inhibited ICC progression, while METTL3 overexpression showed the opposite effect. METTL3 inhibitor STM2457 also showed anti-tumor effect in ICC. Mechanistically, METTL3 transcription was driven by H3K4me3 activation. Upregulation of METTL3 mediated m6A modification of IFIT2 mRNA and accelerated IFIT2 mRNA decay in a YTHDF2-dependent manner, which promoted the development of ICC and lead to poorer prognosis. In summary, our findings revealed that H3K4me3 activation-driven METTL3 transcription promotes ICC progression by YTHDF2-mediated IFIT2 mRNA degradation, suggesting that METTL3 may serve as a potential target for human ICC therapy.
Similar content being viewed by others
Introduction
Intrahepatic cholangiocarcinoma (ICC) is the second most common primary malignant liver cancer, which accounts for ~10% of all such cancers [1, 2]. The prognosis of patients with ICC is poor. Nearly 70% of patients with ICC are already unresectable at the time of diagnosis. Even in patients undergoing curative surgical treatment, the 5-year overall survival (OS) rate is ~30%, and the 5-year recurrence rate is up to 70% [3]. Therefore, effective systemic therapy during the course of the disease is required for the ICC. The combination of gemcitabine and cisplatin is the current first-line therapy for patients with unresectable ICC, but its efficacy remains very limited [4, 5]. Due to the lack of effective treatment and poor prognosis of ICC, understanding the molecular mechanisms underlying ICC development is urgently needed.
Accumulating evidence has revealed that ICC pathogenesis is complicated, which involves epigenetic, genetic, and proteomic alterations [6, 7]. Epigenetic regulation is one of the most common pathways causing gene aberrant expression and facilitating ICC progression [8]. N6-methyadenosine (m6A) modification is the most prevalent mRNA modification in eukaryote [9]. m6A modification is dynamic and reversible, which is regulated by m6A “writer” proteins (Methyltransferases like 3 [METTL3], Methyltransferases like 14 [METTL14] and Wilms tumor 1 associated protein [WTAP]) and “eraser” proteins (Fat-mass and obesity-associated protein [FTO] and Alkylation repair homolog protein 5 [ALKBH5]). In addition, specific “reader” proteins (YTH domain-containing proteins, YTHDF1-3, and YTHDC1-2) can recognize m6A sites and affect RNA process including mRNA stability, decay, splicing, and translation [10, 11]. Aberrant m6A modifications have been found to be involved in the carcinogenesis of a variety of human tumors [12]. However, the mechanism of carcinogenesis influenced by m6A modification dysregulation in ICC remains unclear.
Here, we dissected the reason for the dysregulation of METTL3 in ICC and revealed the regulatory mechanism of the m6A modification mediated by METTL3 in ICC. We also demonstrated that METTL3 regulates IFIT2 expression in an m6A-YTHDF2-dependent manner, and METTL3 may be a novel prognostic predictor and therapeutic target for ICC.
Results
METTL3 expression was elevated in ICC and associated with poorer prognosis
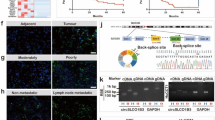
The m6A levels are mainly regulated by m6A writers and erasers. Therefore, we detected mRNA expression of m6A modulators in ICC tissues. The results showed that methyltransferase METTL3 was significantly up-regulated in all ICC data sets, including GEPIA2 dataset (Fig. 1A), GSE107943 (Fig. 1B), and our dataset (Fig. 1C). We then measured the expression level of METTL3 in tumor tissues of 96 ICC patients by immunohistochemistry (IHC) staining (Fig. 1D) and compared the correlation between METTL3 expression and clinical characteristics (Supplementary Table 1). METTL3 expression was positively correlated with tumor size (P < 0.05) (Fig. 1E) and tumor, node, metastasis (TNM) stage (P < 0.05) (Fig. 1F). Kaplan–Meier analysis showed that ICC patients with high METTL3 expression had poorer DFS (P = 0.0025, Fig. 1G) and OS (P = 0.0015, Fig. 1H). Univariate regression analysis illustrated that tumor size, vascular invasion, nerve invasion, TNM stage, lymphatic metastasis, distant metastasis, and METTL3 expression were associated with OS in 96 ICC patients, and Multivariate regression analysis demonstrated that METTL3 expression was an independent predictor for overall survival in patients with ICC (HR = 2.105, 95% CI [1.246–3.557] (Supplementary Table 2 and Fig. 1I)). These results suggest that METTL3 is upregulated in ICC and might be an independent prognostic marker for ICC patients.
A METTL3 expression of ICC tumor (n = 36) and normal tissue (n = 9) in GEPIA2 database. B METTL3 expression of ICC tumor (n = 27) and normal tissue (n = 27) in public dataset GSE107943. C METTL3 expression in ICC tumor (n = 27) and adjacent normal bile duct tissue (n = 8) were measured by real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR). D Representative immunohistochemistry (IHC) staining images of ICC tumors expressing low or high levels of METTL3. E Correlation analysis of METTL3 expression with tumor size (≤5 cm vs. >5 cm). F Correlation analysis of METTL3 expression with tumor, node, metastasis stages (stage I/II vs. III/IV). G Kaplan–Meier survival curves of disease-free survival (DFS) in 96 ICC patients, stratified by METTL3 IHC score (METTL3 low expression, n = 40 vs. METTL3 high expression, n = 56). The P value was calculated using the log-rank test. H Kaplan–Meier survival curves of overall survival (OS) in 96 ICC patients, stratified by METTL3 IHC score (METTL3 low expression, n = 40 vs. METTL3 high expression, n = 56). The P value was calculated using the log-rank test. I Multivariable analyses for overall survival were performed in the ICC cohort. *P < 0.05, ***P < 0.001, according to Student’s t test.
METTL3 transcription is activated by H3K4me3 in ICC
Epigenetic transcriptional activation is an important regulatory pathway of gene transcription. We first analyzed chromatin modification at the transcription start site of METTL3 using the Cistrome Data Browser (http://cistrome.org/). As shown in Fig. 2A, tri-methylation of lysine 4 on histone 3 (H3K4me3) is enriched in the transcription start site of METTL3, indicating that H3K4me3 might regulate the expression of METTL3. SETD1B is a common transcriptional activator with H3K4 tri-methyltransferase activity. Analysis of GEPIA2 data revealed that SETD1B mRNA expression was significantly upregulated in ICC (Fig. 2B) and correlated with METTL3 mRNA expression (Supplementary Fig. 1). The results from our ICC cohort also showed that SETD1B mRNA expression was positively correlated with METTL3 mRNA expression (Fig. 2C). We then knocked down SETD1B mRNA using specific shRNA (Fig. 2D) and found that knockdown of SETD1B mRNA significantly reduced METTL3 mRNA levels (Fig. 2E). The protein levels of METTL3 and H3K4me3 were also reduced (Fig. 2F). Next, we treated ICC cells with CPI-455, a specific KDM5 inhibitor, which can improve the whole level of H3K4me3 modification. The results showed that METTL3 expression significantly increased in a dose-dependent manner (Fig. 2G). Moreover, the chromatin immunoprecipitation assay showed that H3K4me3 signals were enriched in the transcription start site of METTL3, and knockdown of SETD1B could significantly decrease the enrichment of H3K4me3 (Fig. 2H). These results showed that the increase of METTL3 expression may partly be attributed to H3K4me3 enrichment at the transcription start site of METTL3.
A Analysis of H3K4me3 modification in the METTL3 locus in HuCC-T1 cell (datasets from the Cistrome Data Browser). B SETD1B expression in ICC tumors (n = 36) and normal tissues (n = 9) in the GEPIA2 database. C Correlation analysis of METTL3 expression with SETD1B expression in our ICC cohort. D The mRNA levels of SETD1B in SETD1B-KD and control HCCC-9810 cells were confirmed by RT-qPCR. E The mRNA levels of METTL3 in SETD1B-KD and control HCCC-9810 cells were confirmed by RT-qPCR. F The protein levels of METTL3 and H3K4me3 modification after SETD1B silencing in HCCC-9810 cells were confirmed by western blotting. G The mRNA level of METTL3 after CPI-455 treatment for 72 h in HuCC-T1 and HCCC-9810 cells was confirmed by RT-qPCR. H Chromatin immunoprecipitation-qPCR analysis of H3K4me3 enrichment in the METTL3 locus or a flank region with no signal in SETD1B-KD or control HCCC-9810 cells. NT = A flank region with no signal was used as a negative control. The results are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, according to Student’s t test.
Targeting of METTL3 suppresses ICC progression
To determine the biological role of METTL3 in ICC progression, stable METTL3-knockdown ICC cells were established. The knockdown efficiency of METTL3 was verified by qPCR (Fig. 3A) and western blotting (Fig. 3B). Knockdown of METTL3 significantly inhibited ICC cells proliferation (Fig. 3C) and colony formation ability (Fig. 3D and Supplementary Fig. 2A). METTL3 knockdown also significantly promotes ICC cells apoptosis (Fig. 3E) and arrested the cell cycle in S phase (Fig. 3F and Supplementary Fig. 2C). Migration and invasion assays showed that silencing METTL3 significantly inhibited the ability of migration and invasion in ICC cells (Fig. 3G and Supplementary Fig. 2B). Next, METTL3-knockdown HuCC-T1 and HCCC-9810 cells and control cells were subcutaneously injected into mice. The METTL3-knockdown tumors grew more slowly than the tumors in the control group (Fig. 3H and Supplementary Fig. 2D). The tumor volume was calculated every 4 days (Fig. 3I and Supplementary Fig. 2E). The tumor weight was markedly lower in the METTL3-knockdown group compared to the control group (Fig. 3J and Supplementary Fig. 2F). In addition, IHC results showed Ki-67 in METTL3-knockdown tumors was significantly decreased compared with that in the control group (Fig. 3K and Supplementary Fig. 2G). TUNEL assay revealed that more apoptotic cells were found in METTL3-knockdown tumors (Fig. 3L and Supplementary Fig. 2H).
A The mRNA level of METTL3 after METTL3 silencing in HuCC-T1 and HCCC-9810 cells was confirmed by RT-qPCR. B The protein level of METTL3 after METTL3 silencing in HuCC-T1 and HCCC-9810 cells was confirmed by Western blotting. C Cell growth curve of HuCC-T1 and HCCC-9810 cells transfected with METTL3 shRNA or Control. D Colony-forming assays after METTL3 silencing in HuCC-T1 and HCCC-9810 cells. E Apoptosis analysis of HuCC-T1 and HCCC-9810 cells transfected with shControl or shMETTL3. F Cell cycle analysis of HuCC-T1 and HCCC-9810 cells transfected with shControl or shMETTL3. G Cell migration ability and cell invasion ability after shMETTL3-transfection in HuCC-T1 cell. H Xenograft tumors in each group were shown. The mice were sacrificed 28 days post-injection. I Tumor growth curves after the injection of shMETTL3 and Control HuCC-T1 cells. Tumor volume was calculated every 4 days. J Tumor weight of shMETTL3 and Control groups was measured. K Representative IHC staining of Ki67 in tumors with different treatments. L Representative images of TUNEL analysis in tumors with different treatments. The results are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, according to a Student’s t test.
Recently, a novel and selective METTL3 inhibitor, STM2457 has shown therapeutic effect in acute myeloid leukemia [13]. Here, we examine the effect of STM2457 on ICC progression. As expected, STM2457 can inhibit the proliferation (Supplementary Fig. 3A) of ICC cells in a dose-dependent manner. In addition, STM2457 treatment can promote apoptosis of ICC cells (Supplementary Fig. 3B) and arrest the cell cycle of ICC cells in S phase (Supplementary Fig. 3C). Furthermore, STM2457 treatment also inhibited cell invasion and migration of ICC cells in a dose-dependent manner (Supplementary Fig. 3D). These results suggest that therapeutically targeting METTL3 may be a promising treatment for ICC.
METTL3 overexpression promotes ICC progression
To test the effect of METTL3 overexpression on ICC progression, we also established stable METTL3-overexpressing ICC cells. The efficiency of overexpression was verified by qPCR (Fig. 4A) and western blotting (Fig. 4B). Overexpression of METTL3 significantly promotes ICC cells proliferation (Fig. 4C). METTL3 overexpression have mild effect in inhibiting ICC cells apoptosis (Fig. 4D). In addition, METTL3 overexpression reduced the arrest of the cell cycle in S phase (Fig. 4E). Migration and invasion assays showed that METTL3 overexpression significantly promotes the ability of migration and invasion in ICC cells (Fig. 4F). These results suggest that METTL3 is essential in promoting ICC progression.
A The mRNA level of METTL3 after METTL3 overexpression in HuCC-T1 and HCCC-9810 cells was confirmed by RT-qPCR. B The protein level of METTL3 after METTL3 overexpression in HuCC-T1 and HCCC-9810 cells was confirmed by Western blotting. C Cell growth curve of HuCC-T1 and HCCC-9810 cells transfected with METTL3-OE or Vector. D Apoptosis analysis of HuCC-T1 and HCCC-9810 cells transfected with METTL3-OE or Vector. E Cell cycle analysis of HuCC-T1 and HCCC-9810 cells transfected with METTL3-OE or Vector. F Cell migration ability and cell invasion ability of HuCC-T1 and HCCC-9810 cells transfected with METTL3-OE or Vector. The results are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, according to a Student’s t test.
METTL3 facilitates the ICC progression by downregulating IFIT2 expression
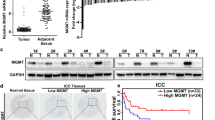
To delineate the molecular mechanism by which METTL3 accelerates ICC progression, RNA-seq in HuCC-T1 cell with or without METTL3 knockdown was performed (Fig. 5A). RNA-seq revealed that 294 transcripts were significantly upregulated upon METTL3 knockdown (Supplementary Fig. 4A), while 488 transcripts were significantly downregulated upon METTL3 knockdown (Supplementary Fig. 4B). We then performed Methylated RNA immunoprecipitation-seq (MeRIP-seq) in HuCC-T1 and HCCC-9810 cells. The motifs of m6A peaks were consistent with the consensus sequence of RRACH (R = G/A, H = A/C/U) (Fig. 5B). There were 28 genes with m6A modification (fold enrichment >4 and P < 0.05) in both HuCC-T1 and HCCC-9810 cells and were differentially expressed (Log2foldchange > 0.5 and P < 0.05) in METTL3 knockdown HuCC-T1 cell (Fig. 5C and Supplementary Fig. 4C). IFIT2 has been reported as a tumor suppressor for several tumor types [14, 15]. Among all the genes, IFIT2 is one of the top up-regulated genes in METTL3 knockdown cells comparing with shControl. The RPKM value of IFIT2 in RNA-seq is shown in Fig. 5D. The mRNA levels of IFIT2 also markedly increased following METTL3 knockdown in another ICC cell line, HCCC-9810 (Fig. 5E). Moreover, the m6A peak in IFIT2 3’UTR is very prominent in MeRIP-seq of both HuCC-T1 and HCCC-9810 cells (Fig. 5F). Then, we measured the mRNA expression of IFIT2 in xenograft tumors. The results showed that the expression of IFIT2 was higher in METTL3 knockdown HuCC-T1 and HCCC-9810 xenograft tumors compared to the control ones (Fig. 5G and Supplementary Fig. 4D). We further measured the protein level of IFIT2 in the tumor tissues of 96 ICC patients by IHC staining (Supplementary Fig. 4E). The expression of IFIT2 in 96 ICC patients was negatively correlated with the expression of METTL3 (Fig. 5H). Kaplan–Meier analysis revealed that ICC patients with low IFIT2 expression had poorer DFS (P = 0.0032, Fig. 5I) and OS (P = 0.0321, Fig. 5J).
A Heatmap showing the expression changes of METTL3-KD and Control HuCC-T1 cell. B The m6A consensus sequence motifs in HuCC-T1 and HCCC-9810 cells was identified. C Scatter plot shows the genes with m6A modification (fold enrichment >4 and P < 0.05) in both HuCC-T1 and HCCC-9810 cells and differentially expressed (Log2foldchange > 0.5 and P < 0.05) in HuCC-T1 cells after METTL3 silencing. D Relative expression of IFIT2 in METTL3-KD and Control HuCC-T1 cells. E RT-qPCR analysis of IFIT2 after METTL3 silencing in HCCC-9810 cells. F Integrative genomics viewer (IGV) plots indicates m6A peaks at IFIT2 mRNAs in MeRIP-seq of ICC cells. The y axis shows the sequence read number, and the blue boxes represent protein-coding exons. G RT-qPCR analysis of IFIT2 mRNA expression in HuCC-T1 xenograft models after METTL3 knockdown or not. H Bar graphs indicate the correlation of METTL3 expression with IFIT2 expression in ICC specimens. I Kaplan–Meier survival curves of DFS in 96 patients with ICC. (IFIT2 low expression, n = 62 vs. IFIT2 high expression, n = 34). The P value was calculated using the log-rank test. HR hazard ratio. J Kaplan–Meier survival curves of OS in 96 patients with ICC. (IFIT2 low expression, n = 62 vs. IFIT2 high expression, n = 34). The P value was calculated using the log-rank test. HR hazard ratio. The results are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, according to Student’s t test.
We then designed two siRNAs targeting IFIT2 and verified the knockdown efficiency by RT-qPCR and western blotting (Figs. 6A, B). Knockdown of IFIT2 have mild effect in promoting cell proliferation of ICC cells (Fig. 6C), and dramatically promoted cell migration, and invasion (Fig. 6D). Then, the expression of IFIT2 was knocked down in METTL3 stable knockdown ICC cells using specific siRNAs (Figs. 6E, F). As expected, knockdown of IFIT2 rescued the proliferation (Fig. 6G), migration, and invasion ability (Fig. 6H) of METTL3 knockdown ICC cells. Our data suggest that METTL3 promotes ICC progression by downregulating the expression of IFIT2.
A The mRNA level of IFIT2 after IFIT2 silencing in HuCC-T1 and HCCC-9810 cells was confirmed by RT-qPCR. B The protein level of IFIT2 after IFIT2 silencing in HuCC-T1 and HCCC-9810 cells was confirmed by western blotting. C Cell growth curve of HuCC-T1 and HCCC-9810 cells transfected with IFIT2 siRNA or Control. D Cell migration ability and cell invasion ability after IFIT2 silencing in HuCC-T1 and HCCC-9810 cells. E The mRNA level of METTL3 and IFIT2 after METTL3 or /and IFIT2 silencing in HuCC-T1 and HCCC-9810 cells was confirmed by RT-qPCR. F The protein level of METTL3 and IFIT2 after METTL3 or /and IFIT2 silencing in HuCC-T1 and HCCC-9810 cells was confirmed by western blotting. G Cell growth curve of HuCC-T1 and HCCC-9810 cells after METTL3 or /and IFIT2 silencing. H Cell migration ability and cell invasion ability in HuCC-T1 and HCCC-9810 cells after METTL3 or /and IFIT2 silencing. The results are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, according to Student’s t test.
METTL3 reduced IFIT2 mRNA stability through an m6A-YTHDF2-dependent pathway
METTL3 usually acts as an oncogene by catalyzing the m6A modification of target genes [16]. To confirm that METTL3 catalyzes the m6A modification of IFIT2, MeRIP-qPCR was performed. The results showed that the m6A modification significantly enriched in IFIT2 mRNA, but decreased in METTL3 knockdown HuCC-T1 and HCCC-9810 cells (Fig. 7A). We then constructed dual luciferase reporter plasmids containing wild-type and mutant 3’ UTR of IFIT2 and relative luciferase activity was compared in both HuCC-T1 and HCCC-9810 cells. The specific mutation position information of the reporter plasmid used is shown in Fig. 7B. Knockdown METTL3 could affect the luciferase activity of the wild-type IFIT2-fused reporter but not the reporter with mutation on the m6A consensus sequences (Fig. 7C). HuCC-T1 and HCCC-9810 cells were treated with a transcription inhibitor (Actinomycin D) for the indicated times. Decelerated mRNA decay was observed upon knockdown of METTL3 (Fig. 7D), while overexpression of METTL3 showed the opposite effect (Fig. 7E). As previously reported [10], YTHDF2 could induce target mRNA degradation by reading m6A modification sites. Therefore, we tested whether YTHDF2 is involved in the regulation of IFIT2 mRNA stability. The mRNA levels of IFIT2 also markedly increased following YTHDF2 knockdown in both HuCC-T1 and HCCC-9810 cells (Fig. 7F). CLIP-seq showed that YTHDF2 can bind to the m6A modification site of IFIT2 in both HuCC-T1 and HCCC-9810 cells (Fig. 7G). Further dual luciferase activity assay also showed that the luciferase activity of wild-type IFIT2-fused reporter was obviously augmented upon YTHDF2 knockdown, but exhibiting no difference in mutation group (Fig. 7H). The actinomycin D assay also showed that knockdown of YTHDF2 could decelerate the mRNA decay of IFIT2 (Fig. 7I), and METTL3 knockdown has no significant effect on decelerating the mRNA decay of IFIT2 in ICC cells with YTHDF2 knockdown (Supplementary Fig. 5). Together, our results showed that METTL3-mediated reduced IFIT2 mRNA stability in a YTHDF2-dependent manner.
A MeRIP-qPCR analysis of m6A enrichment in the IFIT2 locus in METTL3-KD or Control HuCC-T1 and HCCC-9810 cells. NT = A flank region with no signal was used as a negative control. B Schematic representation of mutated (RRAC to RRCC) 3′UTR of psiCHECK2 vector to investigate the roles of m6A in 3′UTR in IFIT2 expression. C Relative luciferase activity of IFIT2 3’UTR with wild-type or mutated m6A sites after METTL3 silencing in HuCC-T1 and HCCC-9810 cells. Renilla luciferase activity was measured and normalized to firefly luciferase activity. D RT-qPCR analysis of IFIT2 after actinomycin D treatment in shMETTL3 or Control HuCC-T1 and HCCC-9810 cells. E RT-qPCR analysis of IFIT2 after actinomycin D treatment in METTL3-OE or Vector HuCC-T1 and HCCC-9810 cells. F The mRNA level of YTHDF2 and IFIT2 after YTHDF2 silencing in HuCC-T1 and HCCC-9810 cells was confirmed by RT-qPCR. G IGV plots indicates YTHDF2 binding at IFIT2 mRNAs. The y axis shows the sequence read number, and the blue boxes represent protein-coding exons. H Relative luciferase activity of IFIT2 3’UTR with wild-type or mutated m6A sites after YTHDF2 silencing in HuCC-T1 and HCCC-9810 cells. Renilla luciferase activity was measured and normalized to firefly luciferase activity. I RT-qPCR analysis of IFIT2 after actinomycin D treatment in shYTHDF2 or Control HuCC-T1 and HCCC-9810 cells. J The schematic diagram for function and mechanism of METTL3 in ICC. The results are presented as mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, according to Student’s t test.
Discussion
The m6A modification is the most prevalent mRNA modification in eukaryote. m6A modification has been reported involving in a variety of human diseases, including cardiovascular disease, metabolic disease, viral infection, and cancer progression [17]. m6A modification has shown potential in cancer by regulating mRNA decay, translation, and processing. The dysregulation of “writer,” “eraser,” and “reader” protein of m6A modification has been found in various human cancers and promotes cancer progression. However, the role and effect of m6A modification in ICC remain unclear. In this study, we first found that the major m6A writer METTL3 was upregulated in ICC. Mechanistically, H3K4me3 activation-driven METTL3 transcription mediated m6A modification of IFIT2 mRNA and accelerated IFIT2 mRNA decay in a YTHDF2-dependent manner. The decreased mRNA levels of IFIT2 promote the progression of ICC and lead to worse clinical prognosis (Fig. 7J).
METTL3 is the major component of the m6A methyltransferase complex. The aberrant transcription of METTL3 has been reported in various cancer types [16]. Previous studies have shown that METTL3 attributes to the oncogenesis of bladder cancer [18], acute myeloid leukemia [19], lung cancer [20], liver cancer [21], breast cancer [22], and gastric cancer [23]. However, the mechanisms for the dysregulation of METTL3 is still unclear. Wang et al. found that METTL3 transcription was activated by H3K27ac modification in gastric cancer [23]. In the present study, we first found that knockdown of tri-methyltransferase decreased the enrichment of H3K4me3 in METTL3 and inhibited the mRNA expression of METTL3. In contrast, KDM5 inhibitors can increase the enrichment of H3K4me3 in METTL3 and increase METTL3 transcription, suggesting that H3K4me3 modification mediated METTL3 transcription in ICC. Loss-of-function and gain-of-function experiments demonstrated the essential role of METTL3 in promoting ICC progression. The METTL3 inhibitor STM2457 also exhibits anti-tumor effect in ICC. Thus, therapeutically targeting METTL3 may be a promising treatment for ICC.
One of the most important functions of m6A modification is epigenetically silencing tumor suppressor genes. Interferon (IFN)-induced protein with tetratricopeptide repeats 2 (IFIT2), also known as ISG54, is a member of the IFN-stimulated genes (ISGs). IFIT2 is a direct response target to type I IFNs (IFN-Is) and plays an important role in the innate immune response [24]. IFIT2 has been identified as a tumor suppressor in several tumor types, such as oral squamous cell carcinoma, breast cancer, gallbladder cancer, and lung cancer [25,26,27,28]. RNA-seq and rescue assays illustrated that IFIT2 was one of the key regulatory targets of METTL3 in ICC. m6A modification relies on reader proteins that binds to m6A modified sites to exert their biological functions. mRNA decay of tumor suppressor genes in cancer progression is mainly mediated by YTHDF2. Wong et al. reported that METTL3 promoted hepatocellular carcinoma carcinogenesis through YTHDF2-dependent post-transcriptional silencing of SOCS2 [21]. Xie et al. reported that YTHDF2 promotes prostate cancer progression by mediating the mRNA degradation of LHPP and NKX3-1 in an m6A-dependent manner and by inducing AKT phosphorylation [29]. In this study, we revealed that METTL3 silenced IFIT2 expression through an m6A-YTHDF2-dependent mechanism. In addition, IFIT2 was shown to be a novel prognostic predictor for ICC.
In conclusion, our study reveals that elevated METTL3 expression correlated with poor prognosis in patients with ICC and plays an oncogenic role in ICC progression. Mechanistically, we found that H3K4me3 activation-driven transcription is the reason for the dysregulation of METTL3 in ICC. METTL3 regulates IFIT2 expression in an m6A-YTHDF2-dependent manner. Therefore, METTL3 may be a novel predictor and therapeutic target for ICC.
Materials and methods
Patients’ specimens
The paraffin-embedded specimens of 96 ICC patients who underwent surgical resection were obtained from the pathology department of the First Affiliated Hospital of Sun Yat-sen University (Guangzhou, China) and used for subsequent immunohistochemical experiments of METTL3 and IFIT2 protein expression. The clinical information of 96 ICC patients were shown in Supplementary Table 3. Another 27 ICC tumor tissues and 8 adjacent normal bile duct tissues for subsequent quantitative real-time PCR of METTL3 or SETD1B mRNA expression were snap-frozen in liquid nitrogen within 30 min after surgical resection. This study was approved by Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University.
Processing of gene expression omnibus (GEO) and the gene expression profiling interactive analysis (GEPIA2) data
The gene expression profiles of GSE107943 were downloaded from the GEO database. The database consisted of 27 pairs of ICC cancer and adjacent non-cancerous tissues samples. The mRNA expression of METTL3 were compared between the two groups.
The GEPIA2 database consisted of 36 ICC cancer and 9 adjacent non-cancerous tissues samples. We compared the mRNA expression of METTL3 and SETD1B between the two groups. The correlation of METTL3 and SETD1B expression in 36 ICC tissues were also analyzed.
Cell culture and transfection
Two human ICC cell lines HuCC-T1 and HCCC-9810 were obtained from Cellcook Co., Ltd. (Guangzhou, China). The ICC cell lines HuCC-T1 and HCCC-9810 were cultured in RPMI 1640 medium (Gibco, USA) supplemented with 10% of fetal bovine serum (FBS, Gibco) in an incubator at 37 °C with 5% CO2.
The transfection of siRNA and shRNA were performed as described previously [30].
Reagents
Targeting human IFIT2 and non-targeting control small interfering RNAs (siRNAs) were purchased from RiboBio Co., Ltd. (Guangzhou, China). The METTL3, YTHDF2 and SETD1B short hairpin RNA (shRNA) sequence were purchased from Sangon Biotech Co., Ltd (Shanghai, China), and then ligated into pLVX-Puro-GFP empty vector to construct shRNA plasmid. The CPI-455 (#S8287) was purchased from Selleck Chemicals (Houston, TX, USA). The STM2457 (#T9060) was purchased from TOPSCIENCE Co. Ltd (Shanghai, China). All sequences for RNA interference and primers sequences used for the experiments were listed in Supplementary Table 4.
Western blotting
Western blotting was performed as described previously [30]. The anti-METTL3 antibody (#ab195352) and anti-Ki67 (#ab156956) were purchased from Abcam. The anti-IFIT2 antibody (#DF8962) was purchased from Affbiotech. The anti-H3K4me3 antibody (#39160) was purchased from Active Motif. The anti-β-actin antibody (#4970) and anti-rabbit IgG, HRP-linked antibody (#7071) was purchased from Cell Signaling Technology. The anti-m6A antibody (#202003) was purchased from Synaptic Systems. The anti-histone H3 antibody (#ab21054) was purchased from Abcam. The anti-YTHDF2 antibody (#24744-1-AP) was purchased from Proteintech.
Dual luciferase reporter assay
The 3’UTR regions of IFIT2 were amplified by PCR from cDNA and cloned into the psicheck-2 vector to construct WT-IFIT2 dual-luciferase reporter plasmid. The 3’UTR fragment which mutated at IFIT2 m6A site (RRACH → RRGCH) was synthesized by Sangon Biotech Co., Ltd and cloned into the psicheck-2 vector to construct Mut-IFIT2 dual-luciferase reporter plasmid.
For dual luciferase reporter assay, the ICC cells with different treatment were transfected in 6-well plates with dual-luciferase reporter plasmid. At 48 h, the luciferase activity was measured by the Dual-luciferase Reporter Assay System (Promega) according to manufacturer’s instructions.
RNA decay assays
ICC cells were seeded in 6-well plate and treated with Actinomycin D (5 μg/ml, #A9415, Sigma–Aldrich, USA) for indicated time. The total RNA was extracted with TRIzol reagent (Life Technologies, USA) and processed RT-qPCR analysis.
Animal experiments
4-week-old female BALB/c nude mice were used for HuCC-T1 tumor xenograft models and 4-week-old female B-NDG® mice (Biocytogen, Beijing, China) were used for HCCC-9810 tumor xenograft models. 1 × 107 HuCC-T1 or HCCC-9810 cells were resuspended in 100 ul PBS with Matrigel (1:1), and injected into the right flank of mice (n = 6/group). The tumor volume was measured every 4 days by caliper. The formula for volume is length × width2/2. After 28 days of implantation, the mice were executed. The xenograft tumors were removed, photographed and weighed. The proliferation index was detected by IHC staining of KI67. The apoptosis cells in tumors were detected by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay. All animals used in the experiment were approved by the First Affiliated Hospital of Sun Yat-sen University ([2020] No. 382).
Immunohistochemical (IHC) staining
This staining assay was performed as described previously [30]. The IHC score was calculated as staining intensity (negative, 0; mild, 1; moderate, 2; severe, 3) multiplying staining area (negative, 0; ≤30%, 1; >30 and ≤60%, 2; >60%, 3) by two experienced pathologists independently. The median scores for METTL3 and IFIT2 were three and four, respectively. For IHC staining of METTL3, IHC score >3 was defined as high METTL3 (n = 56), and IHC score ≤3 was defined as low METTL3 expression (n = 40). For IHC staining of IFIT2, IHC score >3 was defined as high IFIT2 (n = 34), and IHC score ≤3 was defined as low METTL3 expression (n = 62).
ChIP-qPCR
ICC cells were crosslinked in 1% formaldehyde (final concentration) for 10 min at room temperature and followed by 250 mM Glycine quenching. Cell lysates were processed using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology) according to manufacturer’s instructions. The H3K4me3 antibody was used for immunoprecipitation at a dilution factor of 1:1000. ChIP DNA was purified, and subsequent qPCR was conducted using the 2 × ChamQ Universal SYBR qPCR Master Mix (Vazyme). A flank region with no signal was used as a negative control.
RNA-seq
HuCC-T1 cells, infected with lentiviruses expressing shMETTL3-1 and shMETTL3-2, were harvested at 48 h post-infection, followed by RNA extraction using TRIzol solution (Life Technologies). The cDNA library was prepared by Novogene (Beijing, China). The paired-end reads were generated by the Illumina® HiSeq 2500 platform supplied by Novogene. An R package, DESeq, was used to quantify transcription levels and identify differentially expressed genes, using a cut-off of P < 0.05.
MeRIP-seq and MeRIP-qPCR
Total RNA was isolated from indicated HuCC-T1 and HCCC-9810 cells, and the mRNA was further purified using Dynabeads mRNA Purification Kit (61006, Invitrogen, USA). After fragmentation with RNA fragmentation reagent (AM8740, Invitrogen, USA), the anti-m6A antibody was used for immunoprecipitated. Both input and immunoprecipitation RNA samples were then subjected to the sequencing libraries preparation using NEBNext Ultra RNA Library Prep Kit for Illumina and submitted for sequencing on Illumina HiSeq 2500 or MeRIP-qPCR analysis.
CLIP-seq
In short, we induced the covalent cross-link between the protein and the directly bound RNA by ultraviolet irradiation. In this way, RNA-protein crosslinking is achieved. After cross-linking, the RNA is partially hydrolyzed to reduce the bound RNA fragments to a “footprint” size (usually about 30–50 nt), which can be cloned by RNA adaptor ligation and reverse transcription (RT)-PCR amplification. Then sequence these PCR products on a high-throughput platform, which was completed on Illumina Hi-Seq. The RNA-IP performed as described previously [31].
RIP-seq or RIP-qPCR
HuCC-T1 and HCCC-9810 Cells were seeded in a 150-mm dish at a density of 1 × 106 cells/ml. The cells were harvested in lysis buffer. The anti-YTHDF2 antibody was used for immunoprecipitated in a dilution of 1:100. The RNA of input and immunoprecipitated samples were isolated with the TRIzol reagent and subjected to sequencing using Illumina HiSeq 2500 platform supplied by Novogene or qPCR analysis. For qPCR analysis, a flank region with no signal was used as a negative control.
Statistical analysis
The statistical analysis of the data was carried out in SPSS 22.0 software and GraphPad Prism 9 software. The data were expressed as mean ± standard deviation and compared by t test, Wilcoxon test, or Chi square test. DFS and OS were measured by Kaplan–Meier method, and the differences between groups were evaluated by log rank test. The independent predictive factors were determined by the cox proportional hazard model. All the statistical analyses, p < 0.05 were considered statistically significant.
References
Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet.2014;383:2168–79.
Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat Rev Clin Oncol. 2018;15:95–111.
Mavros MN, Economopoulos KP, Alexiou VG, Pawlik TM. Treatment and prognosis for patients with intrahepatic cholangiocarcinoma: systematic review and meta-analysis. JAMA Surg. 2014;149:565–74.
Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med. 2010;362:1273–81.
Shroff RT, Javle MM, Xiao L, Kaseb AO, Varadhachary GR, Wolff RA, et al. Gemcitabine, cisplatin, and nab-Paclitaxel for the treatment of advanced biliary tract cancers: a phase 2 clinical trial. JAMA Oncol. 2019;5:824–30.
Wardell CP, Fujita M, Yamada T, Simbolo M, Fassan M, Karlic R, et al. Genomic characterization of biliary tract cancers identifies driver genes and predisposing mutations. J Hepatol. 2018;68:959–69.
Nepal C, O’Rourke CJ, Oliveira D, Taranta A, Shema S, Gautam P, et al. Genomic perturbations reveal distinct regulatory networks in intrahepatic cholangiocarcinoma. Hepatology.2018;68:949–63.
Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, et al. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov. 2017;7:1116–35.
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell.2012;149:1635–46.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature.2014;505:117–20.
He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176.
Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–17.
Yankova E, Blackaby W, Albertella M, Rak J, De Braekeleer E, Tsagkogeorga G, et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature.2021;593:597–601.
Lai KC, Liu CJ, Lin TJ, Mar AC, Wang HH, Chen CW, et al. Blocking TNF-α inhibits angiogenesis and growth of IFIT2-depleted metastatic oral squamous cell carcinoma cells. Cancer Lett. 2016;370:207–15.
Pidugu VK, Pidugu HB, Wu MM, Liu CJ, Lee TC. Emerging functions of human IFIT proteins in cancer. Front Mol Biosci. 2019;6:148.
Zeng C, Huang W, Li Y, Weng H. Roles of METTL3 in cancer: mechanisms and therapeutic targeting. J Hematol Oncol. 2020;13:117.
Yang C, Hu Y, Zhou B, Bao Y, Li Z, Gong C, et al. The role of m(6)A modification in physiology and disease. Cell Death Dis. 2020;11:960.
Cheng M, Sheng L, Gao Q, Xiong Q, Zhang H, Wu M, et al. The m(6)A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene.2019;38:3667–80.
Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–76.
Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen Z, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18:181.
Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–70.
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z, et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–9.
Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut.2020;69:1193–205.
Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nat Rev Immunol. 2014;14:36–49.
Lai KC, Liu CJ, Chang KW, Lee TC. Depleting IFIT2 mediates atypical PKC signaling to enhance the migration and metastatic activity of oral squamous cell carcinoma cells. Oncogene.2013;32:3686–97.
Koh SY, Moon JY, Unno T, Cho SK. Baicalein suppresses stem cell-like characteristics in radio- and chemoresistant MDA-MB-231 human breast cancer cells through up-regulation of IFIT2. Nutrients. 2019;11:624.
Shen H, Zhan M, Zhang Y, Huang S, Xu S, Huang X, et al. PLZF inhibits proliferation and metastasis of gallbladder cancer by regulating IFIT2. Cell Death Dis. 2018;9:71.
Su W, Xiao W, Chen L, Zhou Q, Zheng X, Ju J, et al. Decreased IFIT2 expression in human non-small-cell lung cancer tissues is associated with cancer progression and poor survival of the patients. Onco Targets Ther. 2019;12:8139–49.
Li J, Xie H, Ying Y, Chen H, Yan H, He L, et al. YTHDF2 mediates the mRNA degradation of the tumor suppressors to induce AKT phosphorylation in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer. 2020;19:152.
Huang CS, Chu J, Zhu XX, Li JH, Huang XT, Cai JP, et al. The C/EBPbeta-LINC01133 axis promotes cell proliferation in pancreatic ductal adenocarcinoma through upregulation of CCNG1. Cancer Lett. 2018;421:63–72.
Moore MJ, Zhang C, Gantman EC, Mele A, Darnell JC, Darnell RB. Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc. 2014;9:263–93.
Acknowledgements
The authors greatly acknowledge the financial support from National Natural Science Foundation of China (81772522, 82002501, 82072644, 81972651, and 31771630), the Guangdong Basic and Applied Basic Research Foundation (2021A1515010123, 2019A1515010096 and 2019A1515010686).
Author information
Authors and Affiliations
Contributions
X-YY conceived the study. X-YY, Q-CX and Y-CT designed the study. Q-CX, Y-CT, Y-HS, SC and Y-QZ performed experiments and analyzed the results. Y-HS, C-SH and X-TH provided and assisted in the collection and analysis of clinical samples. X-YY and WZ supervised and guaranteed the study. X-YY, Q-CX, Y-CT and WZ wrote the manuscript. All authors discussed the results and commented on the manuscript, and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, QC., Tien, YC., Shi, YH. et al. METTL3 promotes intrahepatic cholangiocarcinoma progression by regulating IFIT2 expression in an m6A-YTHDF2-dependent manner. Oncogene 41, 1622–1633 (2022). https://doi.org/10.1038/s41388-022-02185-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-022-02185-1
- Springer Nature Limited
This article is cited by
-
METTL3 promotes osteoblast ribosome biogenesis and alleviates periodontitis
Clinical Epigenetics (2024)
-
N6-methyladenosine-modified circSLCO1B3 promotes intrahepatic cholangiocarcinoma progression via regulating HOXC8 and PD-L1
Journal of Experimental & Clinical Cancer Research (2024)
-
RNA modification-mediated mRNA translation regulation in liver cancer: mechanisms and clinical perspectives
Nature Reviews Gastroenterology & Hepatology (2024)
-
M6A RNA methylation in biliary tract cancer: the function roles and potential therapeutic implications
Cell Death Discovery (2024)
-
METTL3 facilitates renal cell carcinoma progression by PLOD2 m6A-methylation under prolonged hypoxia
Cell Death & Disease (2024)