Abstract
Skeletal muscle, a highly complex muscle type in the eukaryotic system, is characterized by different muscle subtypes and functions associated with specific myosin isoforms. As a result, skeletal muscle is the target of numerous diseases, including distal arthrogryposes (DAs). Clinically, DAs are a distinct disorder characterized by variation in the presence of contractures in two or more distal limb joints without neurological issues. DAs are inherited, and up to 40% of patients with this condition have mutations in genes that encode sarcomeric protein, including myosin heavy chains, troponins, and tropomyosin, as well as myosin binding protein-C (MYBPC). Our research group and others are actively studying the specific role of MYBPC in skeletal muscles. The MYBPC family of proteins plays a critical role in the contraction of striated muscles. More specifically, three paralogs of the MYBPC gene exist, and these are named after their predominant expression in slow-skeletal, fast-skeletal, and cardiac muscle as sMyBP-C, fMyBP-C, and cMyBP-C, respectively, and encoded by the MYBPC1, MYBPC2, and MYBPC3 genes, respectively. Although the physiology of various types of skeletal muscle diseases is well defined, the molecular mechanism underlying the pathological regulation of DAs remains to be elucidated. In this review article, we aim to highlight recent discoveries involving the role of skeletal muscle-specific sMyBP-C and fMyBP-C as well as their expression profile, localization in the sarcomere, and potential role(s) in regulating muscle contractility. Thus, this review provides an overall summary of MYBPC skeletal paralogs, their potential roles in skeletal muscle function, and future research directions.
Similar content being viewed by others
Introduction
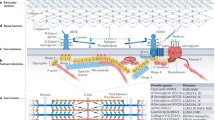
The sarcomere is the smallest functional unit of contraction and the primary component of striated muscle, accounting for over 90% of its protein volume, and acts as a building block of muscles. Proper contraction and relaxation cycles of the sarcomere are critical for the functions of cardiac and skeletal muscles, such as pumping blood out of the heart and maintaining body posture and locomotion, respectively. Thick and thin filament proteins, such as myosin and actin, comprise the two well-defined components in the sarcomere that generate mechanical force by utilizing the energy produced from adenosine triphosphate (ATP) metabolism. However, other sarcomere proteins, including myosin binding protein-C (MyBP-C), are also essential for the steadfast function of striated muscle contractility. Among multiple structural regions of the sarcomere, the C-zone of the A band is the region wherein actin and myosin overlap and form cross-bridges (Fig. 1). MyBP-C, encoded by the MYBPC gene, is specifically localized to the C-zone and regulates the contact of actin and myosin. To date, three MyBP-C paralogs have been identified. Cardiac MyBP-C (cMyBP-C, encoded by MYBPC3) is exclusively expressed in the heart, whereas the other two isoforms, slow and fast MyBP-C (sMyBP-C and fMyBP-C, encoded by MYBPC1 and MYBPC2, respectively), are predominantly expressed in skeletal muscle1. Studies defining the structure and function of MyBP-C in the sarcomere have shed light on the molecular mechanisms underlying the fine-tuning of muscle contraction and relaxation and on the processes and prognoses of cardiomyopathies and skeletal diseases induced by MYBPC gene mutations.
MyBP-C was first discovered in 1971 from rabbit skeletal muscle tissue during myosin filament isolation2. Since then, little progress has been made until the late 1990s, when multiple MYBPC3 mutations were found in patients with hypertrophic cardiomyopathy (HCM)3,4. Over the last two decades, more than 500 MYBPC3 mutations have been found, and the association of MYBPC3 mutations with developing HCM and heart failure (HF) has been intensively studied5,6,7. After the generation of the first knock-in mouse model with the carboxyl-terminal mutation of MYBPC3 developed by the Seidman group in 1999, studies have shown that ablation of cMyBP-C resulted in severe dilated cardiomyopathy8. A similar outcome was observed by the Robbins group in 1999 through the transgenic overexpression of cMyBP-C truncated proteins in mice9. These studies determined the critical role of the carboxyl terminus of cMyBP-C in the development of cardiomyopathies. Additionally, two different knockout (KO) mouse models from two separate laboratories, the Moss laboratory10 and the Carrier laboratory11, independently determined the critical role of cMyBP-C in cardiac function. Moreover, cMyBP-C has multiple phosphorylation sites in its N-terminus, and its binding affinity to the myosin head is distinctly dependent on the phosphorylation status12,13. cMyBP-C phosphorylation has been shown to regulate the speed of muscle contraction via actomyosin interactions14. However, pathological outcomes arise if the level of cMyBP-C phosphorylation is reduced, e.g., cardiomyopathies and HF, implying decreased contractile function15,16. Constitutive cMyBP-C phosphorylation, however, improves cardiac function and has cardioprotective effects after ischemic injury17. Moreover, upon acute cardiac injury, a cleaved N-terminus of cMyBP-C is released into the bloodstream and was thus shown to be a potential biomarker of a compromised heart18,19,20,21. A recent study found that cMyBP-C phosphorylation might regulate calcium transients and sensitivity via direct contact with tropomyosin to enhance binding between calcium and troponin22. Other recent studies have focused on determining the role of the amino terminus region of cMyBP-C23. Overall, these structure-activity studies have further characterized cMyBP-C with a particular focus on the importance of its phosphorylation state.
Compared with those on cMyBP-C, studies on skeletal MyBP-C isoforms (slow and fast MyBP-C) are not as robust, despite the revelations of recent reports showing that mutations in skeletal MyBP-C are associated with inherited skeletal muscle diseases, such as distal arthrogryposis (DA) and lethal congenital contracture syndrome (LCCS)24,25. Nevertheless, multiple phosphorylation sites of sMyBP-C have been identified, and the functional roles of sMyBP-C and fMyBP-C have continued to emerge recently26,27,28. Thus, in this article, we review the basic structure and function of MyBP-C and diseases linked to its mutations. We also discuss possible therapeutic approaches by targeting MyBP-C for the treatment of cardiac and skeletal muscle diseases.
Structure and expression of MYBPC paralogs
MyBP-C is approximately 40 nm long and 3 nm wide with a molecular weight of ~140 kDa29,30,31,32,33. Interestingly, these three paralogs share similar core structural features formed by a linear series of globular domains, including seven immunoglobulin-like and three fibronectin-like domains depicted as C1-C10 from the N-terminus, an additional M-domain linking C1 and C2, and a proline/alanine-rich sequence (PA region) preceding the C1 domain (Fig. 2). Importantly, the cMyBP-C isoform differs from skeletal isoforms with three additional features. First, the cardiac isoform contains an additional immunoglobulin module (C0 domain) at the N-terminus. Second, it has a 28-residue-long loop insertion within the C5 immunoglobulin domain. Finally, it has four phosphorylation sites in the M-domain and a novel phosphorylation site within the PA region14,34. In contrast, the sMyBP-C isoform has one phosphorylation site within the M domain and three phosphorylation sites in the PA region, whereas no phosphorylation site has been reported for fMyBP-C26. The phosphorylation sites of cMyBP-C within the M domain include serine residues at positions 273, 282, 302, and 307 in mice that are the targets of many kinases, including protein kinase A, C and D (PKA, PKC, and PKD, respectively), and calcium/calmodulin-dependent kinase-II (Table 1 and Fig. 2)12,35,36. GSK3β phosphorylates serine 133 within the PA region of cMyBP-C34. In addition to phosphorylation, redox modifications of cMyBP-C have also been reported in different disease models of HF. In contrast to phosphorylation, redox modifications occur through nonenzymatic reactions. These redox modifications include oxidative stress-induced carbonylation, S-nitrosylation, and S-glutathionylation37,38.
The three isoforms of MyBP-C – cardiac, fast-skeletal, and slow-skeletal – all contain seven immunoglobulin domains (Ig), three fibronectin 3 domains (Fn3), an M-domain, and a proline and alanine-rich region. The linker (horizontal line) between immunoglobulin domains C4 and C5 has been reported in both cardiac and slow-skeletal isoforms of MyBP-C. Top: The cardiac isoform (MYBPC3 gene) has an additional Ig domain at the N-terminus, 28 residual inserts in the C5 domain (green vertical line), one phosphorylation site in the proline- and alanine-rich region, and four phosphorylation sites in the M-domain. Middle: The fast-skeletal isoform of MyBP-C (MYBPC2 gene) has no reported phosphorylation sites. Bottom: The slow-skeletal isoform of MyBP-C (MYBPC1 gene) has three phosphorylation sites in the proline- and alanine-rich region and one site in the M-domain.
Despite the suggestive names and their predominant expression patterns that place sMyBP-C and fMyBP-C with skeletal muscles and cMyBP-C exclusively with cardiac tissue39, relatively small, but significant, amounts of sMyBP-C and fMyBP-C are expressed within the heart. In fact, studies have demonstrated that sMyBP-C is expressed in the mammalian heart (i.e., atrium and interatrial septum) and that fMyBP-C is expressed in the heart in murine models of HF40,41. Moreover, even cMyBP-C is expressed in the developing skeletal muscles of chicken embryos42. However, cMyBP-C expression remains restricted to the cardiac muscles of mammalian embryos43.
Although fMyBP-C can be detected at very low levels within slow twitch muscle, it is almost exclusively expressed in fast twitch muscle, specifically within type IIb fibers26,41. Moreover, sMyBP-C is not restricted to slow twitch muscles because it is also abundantly expressed within fast twitch muscles27. Moreover, fMyBP-C and sMyBP-C can be coexpressed within the same muscle types and even coexpressed within the same sarcomere44,45,46,47. Additionally, multiple sMyBP-C variants have been described as expressed in differing amounts in both slow twitch and fast twitch muscles48. In fact, sMyBP-C undergoes a higher level of splicing compared with fMyBP-C and cMyBP-C. At least 14 protein-coding transcripts have been reported in humans and mice that result in sMyBP-C variants with an approximate range between 126 and 132 kDa48,49. Much of this alternative splicing occurs within the first six exons encoding the PA region, although alternative splicing can also occur in regions encoding the M-domain, C7 domain, and carboxyl terminus. Additionally, sMyBP-C variants can be differentially expressed in different muscle groups and even within different myofibers of the same muscle28,48.
Each MyBP-C paralog is known to follow a different spatiotemporal expression pattern. In particular, cMyBP-C is expressed early in embryonic development together with titin and myosin, whereas skeletal muscle paralogs are expressed at later stages of development after the completion of titin and myosin expression, and sMyBP-C expression precedes that of fMyBP-C1,50,51.
Role of MYBPC in cardiac and skeletal muscle function
Based on their phosphorylation status, as suggested previously, sarcomere proteins, including MyBP-C, play key regulatory roles in striated muscle contraction and cardiac hemodynamics52,53. MyBP-C isoforms contain multiple phosphorylation sites targeted by different kinases, such as PKA, PKC, PKD, CaMK, and ribosomal S6 kinase12,18,54,55,56. cMyBP-C is particularly unique because it has a flexible phosphorylation motif that is not present in slow and fast skeletal MyBP-C (Fig. 2). This motif is located at the N-terminal domain and targeted by PKA and CAMK under β-adrenergic stimulation in live myocardium12,57. In sMyBP-C, PKA has been reported to phosphorylate serine 59 and serine 62, whereas PKC phosphorylates serine 83 and threonine 84. Serine 204 is phosphorylated by both PKA and PKC26,58. It has been almost 40 years since the first description of cMyBP-C M domain phosphorylation in intact amphibian myocardium59. Since then, many in vivo and in vitro models and studies have provided more insight into the molecular mechanisms underlying muscle regulation by MyBP-C phosphorylation.
A previous study utilizing skeletal muscle reconstituted with soluble recombinant MyBP-C fragments demonstrated that the presence of the unphosphorylated MyBP-C motif led to a decrease in Ca2+-activated isometric maximal force, an increase in myofilament Ca2+-sensitivity and maximal rigor force and acceleration of the development of rigor force and rigor stiffness60. All these results were eliminated by PKA-dependent phosphorylation of the MyBP-C motif, revealing that the cycling of myosin heads is modulated by the MyBP-C regulatory domain in a phosphorylation-dependent manner. Since the C-terminal domain-dependent anchorage of MyBP-C to the thick filament is not required to obtain such results, it has been suggested that the mechanism of MyBP-C regulation is at least partly independent of a tether60. More recently, this idea was confirmed by a study that used a soluble C1-C2 motif in permeabilized myocytes from wild-type and cMyBP-C-KO mice. The phosphorylation of C1-C2 by PKA reduces the ability of MyBP-C to directly increase myofilament Ca2+ sensitivity. These results demonstrate that the C1-C2:S2 interaction alone is sufficient to disturb the contractility and myofilament Ca2+ sensitivity in a manner independent of a tethering mechanism61. McClellan and colleagues presented an inverse relationship between the percentage of unphosphorylated protein and Ca2+-activated maximal force in which a higher dephosphorylation level corresponds to lower force production62. These findings support a previous study revealing that MyBP-C motif phosphorylation allows myosin heads to extend outward from the thick filament backbone63,64,65,66,67,68. In vitro studies have shown that the MyBP-C N-terminal domain interacts with actin and/or myosin69,70,71. The interaction between the C1-C2 domain and the myosin S2 subdomain (S2) is a phosphorylation-dependent process in which phosphorylated cMyBP-C no longer interacts with S2 but rather moves toward the thin filament to bind actin and thus regulate contractility60,72,73,74,75,76. In addition, the C0 domain of cMyBP-C interacts with myosin regulatory light chain (RLC), which has been verified by multiple in vitro experiments, and HCM-related C0 domain mutations (R35W and K87E) have been shown to reduce the binding affinity of C0 to myosin RLC70. Therefore, the N-terminal domain of cMyBP-C modulates force production and contractile mechanics by regulating the actin-myosin interactions (Fig. 3)77,78,79,80,81.
C-terminal region (C7-C10) is anchored in titin and meromyosin (myosin LMM) and stabilizes the expressed protein. N-terminal (C0-C2) is a functional region that interacts with actin and myosin (S2 and RLC) in a manner dependent on the phosphorylation state and regulates muscle contraction and relaxation.
Cardiomyocyte-specific transgenic mice were generated utilizing the α-myosin promoter to express constitutively active phosphorylated cMyBP-C in which serines 273, 282, and 302 were mutated to aspartic acid17. This chronic phosphorylation causes subtle changes in the sarcomere ultrastructure and an irregular steric arrangement of the thick filament lattice17. In contrast, transgenic mice carrying cMyBP-C, in which the phosphorylation sites are modified to express non-phosphorylatable alanines, show a reduced cardiac contractility, an altered sarcomeric structure and upregulation of transcripts linked to the hypertrophic response18. These studies show how unbalanced levels of MyBP-C phosphorylation can affect the regulation of muscle contraction. In addition, cMyBP-C regulates contractility at the sarcomeric and organ levels, and cMyBP-C phosphorylation is harmonized with the phosphorylation states of other sarcomeric proteins, such as cardiac troponin I82. Similar to cMyBP-C phosphorylation, PKA-mediated sMyBP-C phosphorylation enhances actomyosin interaction by exhibiting accelerated cross-bridge cycling at all levels of Ca2+ activation58. Interestingly, a consensus has not been reached on the regulatory role of cMyBP-C relative to the cross-bridge cycling speed. For example, the results from the rat myocardium show slower cross-bridge cycling, whereas the opposite was observed in the mouse myocardium83,84,85. Despite the discrepancy, possibly from the differences in sample preparation or study methodology, it is possible to deduce that the phosphorylation of cardiac and slow MyBP-C does affect the cross-bridge kinetics. Moreover, the role of fMyBP-C in muscle regulation remains poorly studied. Recently, however, an extensive in vivo and in vitro study was performed with a homozygous Mybpc2-KO mouse model (C2−/−)27. The deletion of fMyBP-C led to a significant reduction in the grip strength, plantar flexor muscle strength, steady-state isometric force during Ca2+ activation, and myofilament calcium sensitivity. These results support previous findings in which the N-terminus of fMyBP-C enhances myofilament Ca2+ sensitivity and force generation86. Furthermore, a structural analysis assessed by small-angle X-ray diffraction of the C2−/− extensor digitorum longus muscle has revealed a greater shift of myosin heads toward actin, less ordered myosin heads, and increased myofilament lattice spacing27. Together, these data demonstrate that fMyBP-C modulates the myofilament Ca2+ sensitivity, actomyosin recruitment, and cross-bridge cycling to match the myofilament contractile output to the demand and that this role is critical for sarcomere integrity, maximal speed, and force generation27.
The regulatory role played by MyBP-C is not restricted to thick filament regulation. A refined study performed with fixed cardiac muscle samples using super-resolution fluorescence microscopy demonstrated that the cMyBP-C N-terminal position is biased toward actin filaments in both active and relaxed muscle76. When a muscle is in its active state, the thin filament sliding process occurs when myosin heads are freed from the thick filament surface and allowed to move toward the thin filament to bind actin87. Notably, it is not necessary for myosin heads to be in an active state to be released from the thick filament88. For example, it has been demonstrated that the N-terminal fragment binds to the thin filament, displaces tropomyosin, locks tropomyosin toward an open conformation, activates cardiac thin filaments, and allows myosin to strongly bind to actin at low Ca2+ levels22. Upon removal of the N-terminal domain or cMyBP-C deletion, cMyBP-C loses its control over actin filament activation and sliding10,77,78,89. Based on its profound impact on actomyosin interactions, the absence of cMyBP-C would result in failed regulation of the bent-back structural configuration (interacting head motif, IHM) and failure to balance the energy-conserving biochemical state (super-relaxed state, SRX) adopted by myosin heads. More specifically, mice lacking MyBP-C exhibit disruption of the myosin population in the SRX state as well as weakened or abolished IHM configurations90,91,92, suggesting that MyBP-C may modulate SRX by stabilizing IHM. Although it has been shown that the cMyBP-C regulatory process is dynamically modulated by cMyBP-C phosphorylation in response to β-adrenergic stimulation, the actual regulatory mechanism remains uncertain90,93. To prevent actomyosin interaction, it has been proposed that the myosin heads fold onto the thick filament and that the MyBP-C N-terminal domain lies along the thick filament surface to stabilize IHM and the very low ATP turnover state of myosin76,94,95,96,97. A study performed with homozygous cMyBP-C-KO mice revealed a decreased myosin population in the SRX state98. In addition, PKA-mediated phosphorylation shifts the biochemical state of the myosin population to a disordered state (from the “OFF” to “ON” state) by reducing cMyBP-C-myosin interactions90,95. Craig and Padron state the following viewpoint: “Enhancement of cardiac contractility by cMyBP-C phosphorylation may result in part from depression of cMyBP-C’s stabilizing effect on the SRX, which coincides with the weakening of the IHM”65,66,67,68.
The distribution of MyBP-C along the different zones of the sarcomere plays an important role in understanding its regulatory control over contractility. By using single-molecule fluorescence imaging of relaxed rat soleus skeletal myofibrils, Nelson and colleagues assessed the kinetics of ATP hydrolysis in the sarcomeric C-zone, D-zone, and P-zone93. The myosin population located at the C-zone, the region in which MyBP-C is abundantly located, is found in the disordered relaxed (DRX) and SRX states and present a longer ATP lifetime compared with other zones that lack MyBP-C. Both the D-zone and P-zone present myosin populations predominantly in the DRX state93. These results confirm the notion that MyBP-C regulates the strong inhibitory state of myosin and can be considered essential for fine-tuning myofilament contractility.
Taken together, the results demonstrate that MyBP-C is a fundamental element in regulating both thin and thick filament activation, actomyosin interaction, cross-bridge kinetics, force generation, and sarcomere integrity. Perhaps it is by selective adaptation that the protein keeps myosin heads in a structurally and biochemically favorable state, according to the muscle’s demands, and thus exerts a dual regulatory effect: inhibition and activation of the striated muscle. Despite significant efforts, we still do not understand the underlying functional and structural roles of MyBP-C or how it modulates myofilament performance.
Regulation of MYBPC and its mutations in muscle diseases
HCM is the most common form of inherited cardiomyopathy, affecting an estimated 1 in 300 individuals around the world99. Accumulated studies over the last two decades have shown that MYBPC3 is the most frequently mutated gene and that it affects approximately 40% of HCM patients. The first two MYBPC3 gene mutations at the C4-C5 linker and C9 domain were reported in patients with HCM by two different research groups in 1995, and since then, over 500 MYBPC3 mutations have been found in HCM patients (Table 1)3,4. Both missense and truncated mutations in the entire MYBPC3 genomic sequence can cause HCM by generating a dominant negative mutation or result in the production of a “poison peptide”100,101. It is a challenge to anticipate mutation penetrance because the binding partner of the mutant protein and additional compounding mutation in the same or different gene can influence the disease profile46. Mybpc3-KO mouse hearts show significant loss of cardiac function and structural changes within 3 days of birth. By adulthood, a significant HCM phenotype has developed in the absence of MYBPC3, and more than 40% of cardiac function is decreased. Although the sarcomeric microstructure is preserved, increased fibrosis and reduced calcium sensitivity are observed in Mybpc3-KO hearts10,102. Both in vivo and in vitro data accumulated from a range of transgenic mouse models and iPSC-derived cardiomyocyte studies have demonstrated that deletion or mutation of MYBPC3 causes HCM by disrupting the regulation of actomyosin, calcium homeostasis, and energy metabolism. In particular, MYBPC3 mutations result in disorganized myosin movement, reduced calcium sensitivity, and elevated energy utilization, all of which contribute to the development of HCM46,103,104. N-terminal cMyBP-C protein (C0-C1 domain) is also cleaved after acute cardiac injury, such as myocardial infarction. Furthermore, this fragment has been shown to exert cytotoxic effects on cardiomyocyte biology, and as such, it can be used as an early indicator of cardiac injury when detected in the circulation105,106.
In contrast to MYBPC3, the pathogenic involvement of skeletal MyBP-C mutations was only recently revealed when MYBPC1 mutations were found in patients with the congenital muscle diseases DA and LCCS. In 2010, Gurnet et al. pioneered the identification of two missense mutations of MYBPC1 in two family members causing the W236R and Y856H amino acid substitutions24. The affected individuals suffer from distal arthrogryposis type 1 (DA1) featuring bilateral clubfoot and camptodactyly with ulnar deviation of the fingers. Since then, more than 10 unique MYBPC1 mutations have been reported to be associated with the development of DA1, DA2 and LCCS25. In a recent publication, Geist et al.107 also showed that the knock-in E248K mutation of MYBPC1 causes severe myopathy characterized by whole-body tremor, kyphosis, functional impairment and muscle atrophy. Similar to cMyBP-C, sMyBP-C also has multiple phosphorylation sites in the N-terminal region targeted by PKA and PKC26. The levels of sMyBP-C phosphorylation are increased after repeated muscle contractions but are decreased in aged and diseased muscles108,109. Moreover, according to a recent report, even transfecting the CRISPR‒Cas9 plasmid with electroporation to knock down 70% of sMyBP-C in adult skeletal muscle causes a significant reduction in peak force generation and the speed of contraction and relaxation. A disoriented sarcomere structure, a stretched Z-disk and a reduced sarcomere length were also observed in this skeletal muscle knockdown study28.
Moreover, two heterozygous compound MYBPC2 mutations (T236I and S255T) were found in Turkish-origin family members with unclassified DA110. Few additional pathogenic MYBPC2 mutations (V307A, F510fs, and A1065V) have been reported, but these remain to be completely characterized (Table 1). Unlike sMyBP-C, no study has reported a phosphorylation site on fMyBP-C. Our recent study, which used the first global Mybpc2-KO mouse model, has demonstrated that fMyBP-C is specifically expressed in fast twitch fibers, type2b and some type2x and is required for maximum force generation in the extensor digitorum longus. Elevated muscle damage and reduced calcium sensitivity are observed in KO muscle concurrent with a disrupted sarcomere microstructure.27
Future directions
The general view holds that cMyBP-C predominates in the heart. However, sMyBP-C is expressed in both slow and fast muscle, and fMyBP-C is enriched in fast skeletal muscle, but low levels also present in slow skeletal muscle and heart muscle. An increasing body of the literature provides a more nuanced picture since the expression profile of MyBP-C paralogs becomes more complex when viewed through the prism of development, adulthood, and development of HF. Sarcomeric proteins play critical, but variable, roles based on their distinct functional properties. cMyBP-C clearly acts as a viscous load by tethering myosin to regulate the rate of cross-bridges and speed of contraction86,111,112,113. Moreover, a significant gap in knowledge accompanies the additional role of skeletal paralogs in slow or fast subtypes of skeletal muscle development and growth as well as their regulatory role(s) in cardiac muscle. We now have a number of novel Mybpc1 and Mybpc2 mouse models, including global and conditional KO mouse lines for sMyBP-C and fMyBP-C, as well as exogenous methods to overexpress these proteins in both skeletal muscle and heart. In addition, we have the ability to carefully interrogate the specific domain regions of sMyBP-C and fMyBP-C by utilizing various in vitro systems to define actomyosin interactions based on comparisons with those of cMyBP-C. Thus, a significant goal of future studies focuses on defining the regulatory roles of sMyBP-C and fMyBP-C in contractility.
Concluding remarks
In summary, the MyBP-C family consists of sarcomeric thick filament proteins essential for normal sarcomere structure and function in cardiac and skeletal muscle. All three MyBP-C paralogs, slow (sMyBP-C), fast (fMyBP-C) and cardiac (cMyBP-C), are encoded by separate genes. Recent studies have established that the cardiomyocyte-specific cMyBP-C is a critical trans-filament protein that acts as a bridge between thick and thin filament proteins via its N’-region. Importantly, mutations in cMyBP-C paralogs are the most common cause of cardiomyopathies. Conversely, sMyBP-C and fMyBP-C are predominantly found in skeletal muscles, and mutations in these two isoforms are known to be associated with skeletal muscle diseases (Fig. 4). To date, a systematic study to determine the specific role of each paralog in skeletal myocytes and cardiomyocytes has yet to be conducted. Several outstanding questions challenge the current dogma regarding the tissue-specific expression and functional relevance of these isoforms; hence, further essential investigations are being conducted to elucidate the potential of modulating these proteins by performing gene therapy as a therapeutic approach.
References
Gautel, M., Furst, D. O., Cocco, A. & Schiaffino, S. Isoform transitions of the myosin binding protein C family in developing human and mouse muscles: lack of isoform transcomplementation in cardiac muscle. Circ. Res. 82, 124–129 (1998).
Starr, R. & Offer, G. Polypeptide chains of intermediate molecular weight in myosin preparations. FEBS Lett. 15, 40–44 (1971).
Watkins, H. et al. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat. Genet. 11, 434–437 (1995).
Bonne, G. et al. Cardiac myosin binding protein-C gene splice acceptor site mutation is associated with familial hypertrophic cardiomyopathy. Nat. Genet. 11, 438–440 (1995).
Carrier, L., Mearini, G., Stathopoulou, K. & Cuello, F. Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene 573, 188–197 (2015).
McNally, E. M., Barefield, D. Y. & Puckelwartz, M. J. The genetic landscape of cardiomyopathy and its role in heart failure. Cell Metab. 21, 174–182 (2015).
van Velzen, H. G. et al. Clinical characteristics and long-term outcome of hypertrophic cardiomyopathy in individuals with a MYBPC3 (Myosin-Binding Protein C) founder mutation. Circ. Cardiovasc. Genet. 10, e001660 (2017).
McConnell, B. K. et al. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J. Clin. Invest. 104, 1235–1244 (1999).
Yang, Q. et al. In vivo modeling of myosin binding protein C familial hypertrophic cardiomyopathy. Circ. Res. 85, 841–847 (1999).
Harris, S. P. et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ. Res. 90, 594–601 (2002).
Carrier, L. et al. Asymmetric septal hypertrophy in heterozygous cMyBP-C null mice. Cardiovasc. Res. 63, 293–304 (2004).
Gautel, M., Zuffardi, O., Freiburg, A. & Labeit, S. Phosphorylation switches specific for the cardiac isoform of myosin binding protein-C: a modulator of cardiac contraction? EMBO J. 14, 1952–1960 (1995).
Previs, M. J. et al. Phosphorylation and calcium antagonistically tune myosin-binding protein C’s structure and function. Proc. Natl Acad. Sci. USA 113, 3239–3244 (2016).
Barefield, D. & Sadayappan, S. Phosphorylation and function of cardiac myosin binding protein-C in health and disease. J. Mol. Cell. Cardiol. 48, 866–875 (2010).
van Dijk, S. J. et al. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation 119, 1473–1483 (2009).
Copeland, O. et al. Analysis of cardiac myosin binding protein-C phosphorylation in human heart muscle. J. Mol. Cell. Cardiol. 49, 1003–1011 (2010).
Sadayappan, S. et al. Cardiac myosin binding protein C phosphorylation is cardioprotective. Proc. Natl Acad. Sci. USA 103, 16918–16923 (2006).
Sadayappan, S. et al. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ. Res. 97, 1156–1163 (2005).
Govindan, S. et al. Cardiac myosin binding protein-C is a potential diagnostic biomarker for myocardial infarction. J. Mol. Cell. Cardiol. 52, 154–164 (2012).
Anand, A. et al. Cardiac myosin-binding protein C is a novel marker of myocardial injury and fibrosis in aortic stenosis. Heart 104, 1101–1108 (2018).
Marber, M. S., Mills, N. L., Morrow, D. A. & Mueller, C. Study Group on Biomarkers of the ESC Association for Acute CardioVascular Care Cardiac myosin-binding protein C as a biomarker of acute myocardial infarction. Eur. Heart J. Acute Cardiovasc. Care 10, 963–965 (2021).
Mun, J. Y. et al. Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc. Natl Acad. Sci. USA 111, 2170–2175 (2014).
Lynch, T. L. T. et al. Amino terminus of cardiac myosin binding protein-C regulates cardiac contractility. J. Mol. Cell. Cardiol. 156, 33–44 (2021).
Gurnett, C. A. et al. Myosin binding protein C1: a novel gene for autosomal dominant distal arthrogryposis type 1. Hum. Mol. Genet. 19, 1165–1173 (2010).
McNamara, J. W. & Sadayappan, S. Skeletal myosin binding protein-C: An increasingly important regulator of striated muscle physiology. Arch. Biochem. Biophys. 660, 121–128 (2018).
Ackermann, M. A. & Kontrogianni-Konstantopoulos, A. Myosin binding protein-C slow is a novel substrate for protein kinase A (PKA) and C (PKC) in skeletal muscle. J. Proteome Res. 10, 4547–4555 (2011).
Song, T. et al. Fast skeletal myosin-binding protein-C regulates fast skeletal muscle contraction. Proc. Natl Acad. Sci. USA 118, e2003596118 (2021).
Geist, J., Ward, C. W. & Kontrogianni-Konstantopoulos, A. Structure before function: myosin binding protein-C slow is a structural protein with regulatory properties. FASEB J. 32, fj201800624R (2018).
Hartzell, H. C. & Sale, W. S. Structure of C protein purified from cardiac muscle. J. Cell Biol. 100, 208–215 (1985).
Heling, L., Geeves, M. A. & Kad, N. M. MyBP-C: one protein to govern them all. J. Muscle Res. Cell Motil. 41, 91–101 (2020).
Offer, G., Moos, C. & Starr, R. A new protein of the thick filaments of vertebrate skeletal myofibrils. Extractions, purification and characterization. J. Mol. Biol. 74, 653–676 (1973).
Weber, F. E., Vaughan, K. T., Reinach, F. C. & Fischman, D. A. Complete sequence of human fast-type and slow-type muscle myosin-binding-protein C (MyBP-C). Differential expression, conserved domain structure and chromosome assignment. Eur. J. Biochem. 216, 661–669 (1993).
Carrier, L. et al. Mapping of a novel gene for familial hypertrophic cardiomyopathy to chromosome 11. Nat. Genet. 4, 311–313 (1993).
Kuster, D. W. et al. GSK3beta phosphorylates newly identified site in the proline-alanine-rich region of cardiac myosin-binding protein C and alters cross-bridge cycling kinetics in human: short communication. Circ. Res. 112, 633–639 (2013).
Jia, W., Shaffer, J. F., Harris, S. P. & Leary, J. A. Identification of novel protein kinase A phosphorylation sites in the M-domain of human and murine cardiac myosin binding protein-C using mass spectrometry analysis. J. Proteome Res. 9, 1843–1853 (2010).
Sadayappan, S. et al. A critical function for Ser-282 in cardiac Myosin binding protein-C phosphorylation and cardiac function. Circ. Res. 109, 141–150 (2011).
Main, A., Fuller, W. & Baillie, G. S. Post-translational regulation of cardiac myosin binding protein-C: a graphical review. Cell. Signal. 76, 109788 (2020).
Figueiredo-Freitas, C. et al. S-Nitrosylation of sarcomeric proteins depresses myofilament Ca2+)sensitivity in intact cardiomyocytes. Antioxid. Redox Signal. 23, 1017–1034 (2015).
Ackermann, M. A. & Kontrogianni-Konstantopoulos, A. Myosin binding protein-C: a regulator of actomyosin interaction in striated muscle. J. Biomed. Biotechnol. 2011, 636403 (2011).
Dhoot, G. K. & Perry, S. V. Expression of slow skeletal myosin binding C-protein in normal adult mammalian heart. J. Muscle Res. Cell Motil. 26, 143–148 (2005).
Lin, B. et al. Cardiac myosin binding protein-C plays no regulatory role in skeletal muscle structure and function. PLoS One 8, e69671 (2013).
Yasuda, M., Koshida, S., Sato, N. & Obinata, T. Complete primary structure of chicken cardiac C-protein (MyBP-C) and its expression in developing striated muscles. J. Mol. Cell. Cardiol. 27, 2275–2286 (1995).
Fougerousse, F. et al. Cardiac myosin binding protein C gene is specifically expressed in heart during murine and human development. Circ. Res. 82, 130–133 (1998).
Reinach, F. C., Masaki, T., Shafiq, S., Obinata, T. & Fischman, D. A. Isoforms of C-protein in adult chicken skeletal muscle: detection with monoclonal antibodies. J. Cell Biol. 95, 78–84 (1982).
Dhoot, G. K., Hales, M. C., Grail, B. M. & Perry, S. V. The isoforms of C protein and their distribution in mammalian skeletal muscle. J. Muscle Res. Cell Motil. 6, 487–505 (1985).
Flashman, E., Redwood, C., Moolman-Smook, J. & Watkins, H. Cardiac myosin binding protein C: its role in physiology and disease. Circ. Res. 94, 1279–1289 (2004).
Reinach, F. C., Masaki, T. & Fischman, D. A. Characterization of the C-protein from posterior latissimus dorsi muscle of the adult chicken: heterogeneity within a single sarcomere. J. Cell Biol. 96, 297–300 (1983).
Ackermann, M. A. & Kontrogianni-Konstantopoulos, A. Myosin binding protein-C slow: a multifaceted family of proteins with a complex expression profile in fast and slow twitch skeletal muscles. Front. Physiol. 4, 391 (2013).
Ackermann, M. A. & Kontrogianni-Konstantopoulos, A. Myosin binding protein-C slow: an intricate subfamily of proteins. J. Biomed. Biotechnol. 2010, 652065 (2010).
Kawashima, M., Kitani, S., Tanaka, T. & Obinata, T. The earliest form of C-protein expressed during striated muscle development is immunologically the same as cardiac-type C-protein. J. Biochem. 99, 1037–1047 (1986).
Kurasawa, M. et al. Differential expression of C-protein isoforms in developing and degenerating mouse striated muscles. Muscle Nerve 22, 196–207 (1999).
Gordon, A. M., Homsher, E. & Regnier, M. Regulation of contraction in striated muscle. Physiol. Rev. 80, 853–924 (2000).
Gupta, M. K. & Robbins, J. Post-translational control of cardiac hemodynamics through myosin binding protein C. Pflug. Arch. 466, 231–236 (2014).
Bardswell, S. C. et al. Distinct sarcomeric substrates are responsible for protein kinase D-mediated regulation of cardiac myofilament Ca2+ sensitivity and cross-bridge cycling. J. Biol. Chem. 285, 5674–5682 (2010).
Cuello, F. et al. Novel role for p90 ribosomal S6 kinase in the regulation of cardiac myofilament phosphorylation. J. Biol. Chem. 286, 5300–5310 (2011).
Mohamed, A. S., Dignam, J. D. & Schlender, K. K. Cardiac myosin-binding protein C (MyBP-C): identification of protein kinase A and protein kinase C phosphorylation sites. Arch. Biochem. Biophys. 358, 313–319 (1998).
Bardswell, S. C., Cuello, F., Kentish, J. C. & Avkiran, M. cMyBP-C as a promiscuous substrate: phosphorylation by non-PKA kinases and its potential significance. J. Muscle Res. Cell Motil. 33, 53–60 (2012).
Robinett, J. C., Hanft, L. M., Geist, J., Kontrogianni-Konstantopoulos, A. & McDonald, K. S. Regulation of myofilament force and loaded shortening by skeletal myosin binding protein C. J. Gen. Physiol. 151, 645–659 (2019).
Hartzell, H. C. Phosphorylation of C-protein in intact amphibian cardiac muscle. Correlation between 32P incorporation and twitch relaxation. J. Gen. Physiol. 83, 563–588 (1984).
Kunst, G. et al. Myosin binding protein C, a phosphorylation-dependent force regulator in muscle that controls the attachment of myosin heads by its interaction with myosin S2. Circ. Res. 86, 51–58 (2000).
Harris, S. P., Rostkova, E., Gautel, M. & Moss, R. L. Binding of myosin binding protein-C to myosin subfragment S2 affects contractility independent of a tether mechanism. Circ. Res. 95, 930–936 (2004).
McClellan, G., Kulikovskaya, I. & Winegrad, S. Changes in cardiac contractility related to calcium-mediated changes in phosphorylation of myosin-binding protein C. Biophys. J. 81, 1083–1092 (2001).
Weisberg, A. & Winegrad, S. Alteration of myosin cross bridges by phosphorylation of myosin-binding protein C in cardiac muscle. Proc. Natl Acad. Sci. USA 93, 8999–9003 (1996).
Weisberg, A. & Winegrad, S. Relation between crossbridge structure and actomyosin ATPase activity in rat heart. Circ. Res. 83, 60–72 (1998).
Caremani, M. et al. Inotropic interventions do not change the resting state of myosin motors during cardiac diastole. J. Gen. Physiol. 151, 53–65 (2019).
Craig, R. & Padron, R. Structural basis of the super- and hyper-relaxed states of myosin II. J. Gen. Physiol. 154, e202113012 (2022).
Irving, T. C. & Craig, R. Getting into the thick (and thin) of it. J. Gen. Physiol. 151, 610–613 (2019).
Kensler, R. W., Craig, R. & Moss, R. L. Phosphorylation of cardiac myosin binding protein C releases myosin heads from the surface of cardiac thick filaments. Proc. Natl Acad. Sci. USA 114, E1355–E1364 (2017).
Mun, J. Y. et al. Electron microscopy and 3D reconstruction of F-actin decorated with cardiac myosin-binding protein C (cMyBP-C). J. Mol. Biol. 410, 214–225 (2011).
Ratti, J., Rostkova, E., Gautel, M. & Pfuhl, M. Structure and interactions of myosin-binding protein C domain C0: cardiac-specific regulation of myosin at its neck? J. Biol. Chem. 286, 12650–12658 (2011).
Razumova, M. V. et al. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: Evidence for long-lived cross-bridges. J. Biol. Chem. 281, 35846–35854 (2006).
Gruen, M. & Gautel, M. Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J. Mol. Biol. 286, 933–949 (1999).
Gruen, M., Prinz, H. & Gautel, M. cAPK-phosphorylation controls the interaction of the regulatory domain of cardiac myosin binding protein C with myosin-S2 in an on-off fashion. FEBS Lett. 453, 254–259 (1999).
Kulikovskaya, I., McClellan, G., Flavigny, J., Carrier, L. & Winegrad, S. Effect of MyBP-C binding to actin on contractility in heart muscle. J. Gen. Physiol. 122, 761–774 (2003).
Levine, R., Weisberg, A., Kulikovskaya, I., McClellan, G. & Winegrad, S. Multiple structures of thick filaments in resting cardiac muscle and their influence on cross-bridge interactions. Biophys. J. 81, 1070–1082 (2001).
Rahmanseresht, S. et al. The N terminus of myosin-binding protein C extends toward actin filaments in intact cardiac muscle. J. Gen. Physiol. 153, e202012726 (2021).
Previs, M. J., Michalek, A. J. & Warshaw, D. M. Molecular modulation of actomyosin function by cardiac myosin-binding protein C. Pflug. Arch. 466, 439–444 (2014).
Previs, M. J., Beck Previs, S., Gulick, J., Robbins, J. & Warshaw, D. M. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science 337, 1215–1218 (2012).
Moss, R. L., Fitzsimons, D. P. & Ralphe, J. C. Cardiac MyBP-C regulates the rate and force of contraction in mammalian myocardium. Circ. Res. 116, 183–192 (2015).
Oakley, C. E., Chamoun, J., Brown, L. J. & Hambly, B. D. Myosin binding protein-C: enigmatic regulator of cardiac contraction. Int J. Biochem. Cell Biol. 39, 2161–2166 (2007).
Pfuhl, M. & Gautel, M. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): who does what, with what, and to whom? J. Muscle Res. Cell Motil. 33, 83–94 (2012).
Yang, Q. et al. PKA-dependent phosphorylation of cardiac myosin binding protein C in transgenic mice. Cardiovasc. Res 51, 80–88 (2001).
Hanft, L. M. et al. Molecule specific effects of PKA-mediated phosphorylation on rat isolated heart and cardiac myofibrillar function. Arch. Biochem. Biophys. 601, 22–31 (2016).
Stelzer, J. E., Patel, J. R. & Moss, R. L. Protein kinase A-mediated acceleration of the stretch activation response in murine skinned myocardium is eliminated by ablation of cMyBP-C. Circ. Res. 99, 884–890 (2006).
Hanft, L. M. & McDonald, K. S. Sarcomere length dependence of power output is increased after PKA treatment in rat cardiac myocytes. Am. J. Physiol. Heart Circ. Physiol. 296, H1524–H1531 (2009).
Lin, B. L. et al. Skeletal myosin binding protein-C isoforms regulate thin filament activity in a Ca(2+)-dependent manner. Sci. Rep. 8, 2604 (2018).
Kampourakis, T., Yan, Z., Gautel, M., Sun, Y. B. & Irving, M. Myosin binding protein-C activates thin filaments and inhibits thick filaments in heart muscle cells. Proc. Natl Acad. Sci. USA 111, 18763–18768 (2014).
Caremani, M. et al. Low temperature traps myosin motors of mammalian muscle in a refractory state that prevents activation. J. Gen. Physiol. 151, 1272–1286 (2019).
Napierski, N. C. et al. A novel “cut and paste” method for in situ replacement of cMyBP-C reveals a new role for cMyBP-C in the regulation of contractile oscillations. Circ. Res. 126, 737–749 (2020).
McNamara, J. W., Singh, R. R. & Sadayappan, S. Cardiac myosin binding protein-C phosphorylation regulates the super-relaxed state of myosin. Proc. Natl Acad. Sci. USA 116, 11731–11736 (2019).
Woodhead, J. L. et al. Atomic model of a myosin filament in the relaxed state. Nature 436, 1195–1199 (2005).
Zoghbi, M. E., Woodhead, J. L., Moss, R. L. & Craig, R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc. Natl Acad. Sci. USA 105, 2386–2390 (2008).
Nelson, S. R., Li, A., Beck-Previs, S., Kennedy, G. G. & Warshaw, D. M. Imaging ATP consumption in resting skeletal muscle: one molecule at a time. Biophys. J. 119, 1050–1055 (2020).
Brunello, E. et al. Myosin filament-based regulation of the dynamics of contraction in heart muscle. Proc. Natl Acad. Sci. USA 117, 8177–8186 (2020).
Nag, S. et al. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol. 24, 525–533 (2017).
Sarkar, S. S. et al. The hypertrophic cardiomyopathy mutations R403Q and R663H increase the number of myosin heads available to interact with actin. Sci. Adv. 6, eaax0069 (2020).
Trivedi, D. V., Adhikari, A. S., Sarkar, S. S., Ruppel, K. M. & Spudich, J. A. Hypertrophic cardiomyopathy and the myosin mesa: viewing an old disease in a new light. Biophys. Rev. 10, 27–48 (2018).
McNamara, J. W. et al. Ablation of cardiac myosin binding protein-C disrupts the super-relaxed state of myosin in murine cardiomyocytes. J. Mol. Cell. Cardiol. 94, 65–71 (2016).
Semsarian, C., Ingles, J., Maron, M. S. & Maron, B. J. New perspectives on the prevalence of hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 65, 1249–1254 (2015).
Ross, S. B., Bagnall, R. D., Ingles, J., Van Tintelen, J. P. & Semsarian, C. Burden of recurrent and ancestral mutations in families with hypertrophic cardiomyopathy. Circ. Cardiovasc. Genet. 10, e001671 (2017).
Kuster, D. W. et al. A hypertrophic cardiomyopathy-associated MYBPC3 mutation common in populations of South Asian descent causes contractile dysfunction. J. Biol. Chem. 290, 5855–5867 (2015).
McConnell, B. K. et al. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J. Clin. Invest. 104, 1771 (1999).
Helms, A. S. et al. Effects of MYBPC3 loss-of-function mutations preceding hypertrophic cardiomyopathy. JCI Insight 5, e133782 (2020).
Toepfer, C. N. et al. Hypertrophic cardiomyopathy mutations in MYBPC3 dysregulate myosin. Sci. Transl. Med. 11, eaat1199 (2019).
Lynch, T. L. & Sadayappan, S. Surviving the infarct: a profile of cardiac myosin binding protein-C pathogenicity, diagnostic utility, and proteomics in the ischemic myocardium. Proteom. Clin. Appl. 8, 569–577 (2014).
Govindan, S. et al. Pathogenic properties of the N-terminal region of cardiac myosin binding protein-C in vitro. J. Muscle Res. Cell Motil. 33, 17–30 (2012).
Geist Hauserman, J. et al. Sarcomeric deficits underlie MYBPC1-associated myopathy with myogenic tremor. JCI Insight 6, e147612 (2021).
Ackermann, M. A., Kerr, J. P., King, B., C, W. W. & Kontrogianni-Konstantopoulos, A. The phosphorylation profile of myosin binding protein-C slow is dynamically regulated in slow-twitch muscles in health and disease. Sci. Rep. 5, 12637 (2015).
Ackermann, M. A., Ward, C. W., Gurnett, C. & Kontrogianni-Konstantopoulos, A. Myosin binding protein-C slow phosphorylation is altered in duchenne dystrophy and arthrogryposis myopathy in fast-twitch skeletal muscles. Sci. Rep. 5, 13235 (2015).
Bayram, Y. et al. Molecular etiology of arthrogryposis in multiple families of mostly Turkish origin. J. Clin. Invest 126, 762–778 (2016).
Li, A. et al. Skeletal MyBP-C isoforms tune the molecular contractility of divergent skeletal muscle systems. Proc. Natl Acad. Sci. USA 116, 21882–21892 (2019).
Chen, Z., Li, X. Y., Guo, P. & Wang, D. L. MYBPC2 and MYL1 as Significant Gene Markers for Rhabdomyosarcoma. Technol. Cancer Res. Treat. 20, 1–15 (2021).
Ducret, V. et al. Transcriptomic analysis of the trade-off between endurance and burst-performance in the frog Xenopus allofraseri. BMC Genomics 22, 204 (2021).
Li, X. et al. Two novel mutations in myosin binding protein C slow causing distal arthrogryposis type 2 in two large Han Chinese families may suggest important functional role of immunoglobulin domain C2. PLoS One 10, e0117158 (2015).
Stavusis, J. et al. Novel mutations in MYBPC1 are associated with myogenic tremor and mild myopathy. Ann. Neurol. 86, 129–142 (2019).
Ha, K. et al. MYBPC1 mutations impair skeletal muscle function in zebrafish models of arthrogryposis. Hum. Mol. Genet. 22, 4967–4977 (2013).
Li, M., Andersson-Lendahl, M., Sejersen, T. & Arner, A. Knockdown of fast skeletal myosin-binding protein C in zebrafish results in a severe skeletal myopathy. J. Gen. Physiol. 147, 309–322 (2016).
Acknowledgements
S.S. has received support from the National Institutes of Health grants R01 AR079435, R01 AR079477, R01 AR078001, R01 HL105826, R01 HL130356, R38 HL155775 and R01 HL143490, the American Heart Association 2019 Institutional Undergraduate Student (19UFEL34380251) and Transformation (19TPA34830084) awards, the PLN Foundation (PLN crazy idea) and the Leducq Foundation (Transatlantic Network 18CVD01, PLN-CURE). Dr. Pinto is supported by the National Institutes of Health grants RO1 HL160966 and R21 AR077802. O.K. has received support from the National Institutes of Health grants R01 HL148598 and R01 AI159078 and an American Heart Association Career Development Award (18CDA34110117). T.S. (19POST34380448) and M.L.V. (2021 AHAPRE216237) were supported by American Heart Association Fellowship training grants.
Author information
Authors and Affiliations
Contributions
T.S., M.O., M.L.-V., C.G., O.K., J.R.P., and S.S. drafted the manuscript and figures. T.S., J.R.P. and S.S. edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.S. performs consulting and collaborative research studies with the Leducq Foundation (CURE-PLAN), Red Saree Inc., Greater Cincinnati Tamil Sangam, Novo Nordisk, Pfizer, AavantioBio, AstraZeneca, MyoKardia, Merck and Amgen, but such work is unrelated to the content of this article.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Song, T., Landim-Vieira, M., Ozdemir, M. et al. Etiology of genetic muscle disorders induced by mutations in fast and slow skeletal MyBP-C paralogs. Exp Mol Med 55, 502–509 (2023). https://doi.org/10.1038/s12276-023-00953-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s12276-023-00953-x
- Springer Nature Limited
This article is cited by
-
Fast myosin binding protein C knockout in skeletal muscle alters length-dependent activation and myofilament structure
Communications Biology (2024)
-
Myosin-binding protein C regulates the sarcomere lattice and stabilizes the OFF states of myosin heads
Nature Communications (2024)