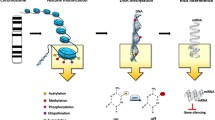
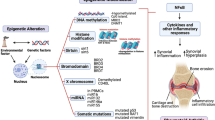
Key Points
-
Epigenetic mechanisms are essential for immune cell differentiation and function, including the correct activation of B cells and T cells and inflammatory processes
-
The dysregulation of epigenetic mechanisms in genetically predisposed individuals is associated with inflammatory rheumatic diseases
-
Epigenome-wide association studies in genetically complex inflammatory rheumatic diseases have identified substantial correlations between epigenetic mechanisms and disease activity and severity
-
Epigenetic dysregulation contributes to the clinical manifestations of monogenic autoinflammatory syndromes and can be used as a biomarker of response to treatment
-
The systematic use of epigenomic screening will help to classify and identify novel biomarkers for personalized management of patients with inflammatory rheumatic diseases
-
New inhibitors of epigenetic enzymes or upstream enzymes that are linked to the epigenetic control of immune function are likely to be tested in clinical trials for disease management
Abstract
Over the past decade, awareness of the importance of epigenetic alterations in the pathogenesis of rheumatic diseases has grown in parallel with a general recognition of the fundamental role of epigenetics in the regulation of gene expression. Large-scale efforts to generate genome-wide maps of epigenetic modifications in different cell types, as well as in physiological and pathological contexts, illustrate the increasing recognition of the relevance of epigenetics. To date, although several reports have demonstrated the occurrence of epigenetic alterations in a wide range of inflammatory rheumatic conditions, epigenomic information is rarely used in a clinical setting. By contrast, several epigenetic biomarkers and treatments are currently in use for personalized therapies in patients with cancer. This Review highlights advances from the past 5 years in the field of epigenetics and their application to inflammatory rheumatic diseases, delineating the future lines of development for a rational use of epigenetic information in clinical settings and in personalized medicine. These advances include the identification of epipolymorphisms associated with clinical outcomes, DNA methylation as a contributor to disease susceptibility in rheumatic conditions, the discovery of novel epigenetic mechanisms that modulate disease susceptibility and the development of new epigenetic therapies.


Similar content being viewed by others
References
Geissmann, F. et al. Development of monocytes, macrophages, and dendritic cells. Science 327, 656–661 (2010).
Álvarez-Errico, D., Vento-Tormo, R., Sieweke, M. & Ballestar, E. Epigenetic control of myeloid cell differentiation, identity and function. Nat. Rev. Immunol. 15, 7–17 (2014).
Ballestar, E. Epigenetic alterations in autoimmune rheumatic diseases. Nat. Rev. Rheumatol. 7, 263–271 (2011).
Rakyan, V. K., Down, T. A., Balding, D. J. & Beck, S. Epigenome-wide association studies for common human diseases. Nat. Rev. Genet. 12, 529–541 (2011).
Javierre, B. M. et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell 167, 1369–1384.e19 (2016).
Feil, R. & Fraga, M. F. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 13, 97–109 (2012).
Javierre, B. M. et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 20, 170–179 (2010).
Viatte, S., Plant, D. & Raychaudhuri, S. Genetics and epigenetics of rheumatoid arthritis. Nat. Rev. Rheumatol. 9, 141–153 (2013).
Vento-Tormo, R. et al. DNA demethylation of inflammasome-associated genes is enhanced in patients with cryopyrin-associated periodic syndromes. J. Allergy Clin. Immunol. 139, 202–211.e6 (2017).
Avgustinova, A. & Aznar Benitah, S. Epigenetic control of adult stem cell function. Nat. Rev. Mol. Cell Biol. 17, 642–658 (2016).
Sun, D. et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell 14, 673–688 (2014).
Farlik, M. et al. DNA methylation dynamics of human hematopoietic stem cell differentiation. Cell Stem Cell 19, 808–822 (2016).
Sellars, M. et al. Regulation of DNA methylation dictates CD4 expression during the development of helper and cytotoxic T cell lineages. Nat. Immunol. 16, 746–754 (2015).
Lai, A. Y. et al. DNA methylation profiling in human B cells reveals immune regulatory elements and epigenetic plasticity at Alu elements during B cell activation. Genome Res. 23, 2030–2041 (2013).
Ichiyama, K. et al. The methylcytosine dioxygenase TET2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity 42, 613–626 (2015).
Vento-Tormo, R. et al. IL-4 orchestrates STAT6-mediated DNA demethylation leading to dendritic cell differentiation. Genome Biol. 17, 4 (2016).
de la Rica, L. et al. PU.1 target genes undergo TET2-coupled demethylation and DNMT3B-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 14, R99 (2013).
Cheng, S.-C. et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684 (2014).
Saeed, S. et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345, 1251086 (2014).
Novakovic, B. et al. β-Glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167, 1354–1368.e14 (2016).
Hedl, M., Li, J., Cho, J. H. & Abraham, C. Chronic stimulation of NOD2 mediates tolerance to bacterial products. Proc. Natl Acad. Sci. USA 104, 19440–19445 (2007).
Watanabe, T. et al. Muramyl dipeptide activation of nucleotide-binding oligomerization domain 2 protects mice from experimental colitis. J. Clin. Invest. 118, 545–559 (2008).
Ellinghaus, D. et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat. Genet. 48, 510–518 (2016).
Duerr, R. H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006).
Parkes, M., Cortes, A., van Heel, D. A. & Brown, M. A. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat. Rev. Genet. 14, 661–673 (2013).
Jin, J. et al. Epigenetic regulation of the expression of IL12 and IL23 and autoimmune inflammation by the deubiquitinase Trabid. Nat. Immunol. 17, 259–268 (2016).
Lu, Q., Chang, C. C. & Richardson, B. C. (eds) Epigenetics and Dermatology (Academic Press, 2015).
Lei, W. et al. Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand. J. Rheumatol. 38, 369–374 (2009).
Houseman, E. A. et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86 (2012).
Liu, Y. et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 31, 142–147 (2013).
Ai, R. et al. Joint-specific DNA methylation and transcriptome signatures in rheumatoid arthritis identify distinct pathogenic processes. Nat. Commun. 7, 11849 (2016).
Frank-Bertoncelj, M. et al. Epigenetically-driven anatomical diversity of synovial fibroblasts guides joint-specific fibroblast functions. Nat. Commun. 8, 14852 (2017).
Spreafico, R. et al. Epipolymorphisms associated with the clinical outcome of autoimmune arthritis affect CD4+ T cell activation pathways. Proc. Natl Acad. Sci. USA 113, 13845–13850 (2016).
Meyer, B. et al. DNA methylation at IL32 in juvenile idiopathic arthritis. Sci. Rep. 5, 11063 (2015).
Kim, S. H., Han, S. Y., Azam, T., Yoon, D. Y. & Dinarello, C. A. Interleukin-32: a cytokine and inducer of TNFα. Immunity 22, 131–142 (2005).
Heinhuis, B. et al. Inflammation-dependent secretion and splicing of IL-32γ in rheumatoid arthritis. Proc. Natl Acad. Sci. USA 108, 4962–4967 (2011).
Coit, P. et al. Epigenome profiling reveals significant DNA demethylation of interferon signature genes in lupus neutrophils. J. Autoimmun. 58, 59–66 (2015).
Elkon, K. B. & Stone, V. V. Type I interferon and systemic lupus erythematosus. J. Interferon Cytokine Res. 31, 803–812 (2011).
Coit, P. et al. Epigenetic reprogramming in naïve CD4+ T cells favoring T cell activation and non-Th1 effector T cell immune response as an early event in lupus flares. Arthritis Rheumatol. 68, 2200–2209 (2016).
Chan, R. W. Y. et al. Plasma DNA aberrations in systemic lupus erythematosus revealed by genomic and methylomic sequencing. Proc. Natl Acad. Sci. USA 111, E5302–E5311 (2014).
Lun, F. M. F. et al. Noninvasive prenatal methylomic analysis by genomewide bisulfite sequencing of maternal plasma DNA. Clin. Chem. 59, 1583–1594 (2013).
Brown, M. A. et al. The effect of HLA-DR genes on susceptibility to and severity of ankylosing spondylitis. Arthritis Rheum. 41, 460–465 (1998).
Wellcome Trust Case Control Consortium et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 (2007).
Isakov, N. & Altman, A. PKCθ-mediated signal delivery from the TCR/CD28 surface receptors. Front. Immunol. 3, 273 (2012).
Roberts, A. R. et al. An ankylosing spondylitis-associated genetic variant in the IL23R-IL12RB2 intergenic region modulates enhancer activity and is associated with increased Th1-cell differentiation. Ann. Rheum. Dis. 75, 2150–2156 (2016).
Ong, C. & Corces, V. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 12, 283–293 (2011).
Aksentijevich, I. et al. Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell 90, 797–807 (1997).
Centola, M. et al. The gene for familial Mediterranean fever, MEFV, is expressed in early leukocyte development and is regulated in response to inflammatory mediators. Blood 95, 3223–3231 (2000).
Xu, H. et al. Innate immune sensing of bacterial modifications of Rho GTPases by the pyrin inflammasome. Nature 513, 237–241 (2014).
Yu, J.-W. et al. Cryopyrin and pyrin activate caspase-1, but not NF-κB, via ASC oligomerization. Cell Death Differ. 13, 236–249 (2006).
Lachmann, H. J. et al. Clinical and subclinical inflammation in patients with familial Mediterranean fever and in heterozygous carriers of MEFV mutations. Rheumatology (Oxford) 45, 746–750 (2006).
Gershoni-Baruch, R. et al. The contribution of genotypes at the MEFV and SAA1 loci to amyloidosis and disease severity in patients with familial Mediterranean fever. Arthritis Rheum. 48, 1149–1155 (2003).
Aypar, E. et al. Th1 polarization in familial Mediterranean fever. J. Rheumatol. 30, 2011–2013 (2003).
Ben-Zvi, I., Brandt, B., Berkun, Y., Lidar, M. & Livneh, A. The relative contribution of environmental and genetic factors to phenotypic variation in familial Mediterranean fever (FMF). Gene 491, 260–263 (2012).
Kirectepe, A. K. et al. Analysis of MEFV exon methylation and expression patterns in familial Mediterranean fever. BMC Med. Genet. 12, 105 (2011).
Mortimer, L., Moreau, F., MacDonald, J. A. & Chadee, K. NLRP3 inflammasome inhibition is disrupted in a group of auto-inflammatory disease CAPS mutations. Nat. Immunol. 17, 1176–1186 (2016).
Zhou, Q. et al. Brief report: cryopyrin-associated periodic syndrome caused by a myeloid-restricted somatic NLRP3 mutation. Arthritis Rheumatol. 67, 2482–2486 (2015).
Lamkanfi, M. & Dixit, V. M. Mechanisms and functions of inflammasomes. Cell 157, 1013–1022 (2014).
Man, S. M. & Kanneganti, T. D. Regulation of inflammasome activation. Immunol. Rev. 265, 6–21 (2015).
Takeuchi, O. & Akira, S. Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010).
Martinon, F., Burns, K. & Tschopp, J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 (2002).
Wang, L. et al. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-κB and caspase-1-dependent cytokine processing. J. Biol. Chem. 277, 29874–29880 (2002).
Dinarello, C. A. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 27, 519–550 (2009).
He, Y., Hara, H. & Núñez, G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem. Sci. 41, 1012–1021 (2016).
Aksentijevich, I. et al. The clinical continuum of cryopyrinopathies: novel CIAS1 mutations in North American patients and a new cryopyrin model. Arthritis Rheum. 56, 1273–1285 (2007).
Aubert, P. et al. Homeostatic tissue responses in skin biopsies from NOMID patients with constitutive overproduction of IL-1β. PLoS ONE 7, e49408 (2012).
Angiolilli, C. et al. Histone deacetylase 3 regulates the inflammatory gene expression programme of rheumatoid arthritis fibroblast-like synoviocytes. Ann. Rheum. Dis. 76, 277–285 (2017).
Toussirot, E. et al. Imbalance between HAT and HDAC activities in the PBMCs of patients with ankylosing spondylitis or rheumatoid arthritis and influence of HDAC inhibitors on TNFα production. PLoS ONE 8, e70939 (2013).
Angiolilli, C. et al. Inflammatory cytokines epigenetically regulate rheumatoid arthritis fibroblast-like synoviocyte activation by suppressing HDAC5 expression. Ann. Rheum. Dis. 75, 430–438 (2016).
Hu, N. et al. Abnormal histone modification patterns in lupus CD4+ T cells. J. Rheumatol. 35, 804–810 (2008).
Leoni, F. et al. The antitumor histone deacetylase inhibitor suberoylanilide hydroxamic acid exhibits antiinflammatory properties via suppression of cytokines. Proc. Natl Acad. Sci. USA 99, 2995–3000 (2002).
Joosten, L. A. B., Leoni, F., Meghji, S. & Mascagni, P. Inhibition of HDAC activity by ITF2357 ameliorates joint inflammation and prevents cartilage and bone destruction in experimental arthritis. Mol. Med. 17, 391–396 (2011).
Nishida, K. et al. Histone deacetylase inhibitor suppression of autoantibody-mediated arthritis in mice via regulation of p16INK4a and p21 WAF1/Cip1 expression. Arthritis Rheum. 50, 3365–3376 (2004).
Vojinovic, J. et al. Safety and efficacy of an oral histone deacetylase inhibitor in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum. 63, 1452–1458 (2011).
Grabiec, A. M., Korchynskyi, O., Tak, P. P. & Reedquist, K. A. Histone deacetylase inhibitors suppress rheumatoid arthritis fibroblast-like synoviocyte and macrophage IL-6 production by accelerating mRNA decay. Ann. Rheum. Dis. 71, 424–431 (2012).
Hsieh, I.-N. et al. Preclinical anti-arthritic study and pharmacokinetic properties of a potent histone deacetylase inhibitor MPT0G009. Cell Death Dis. 5, e1166 (2014).
Shu, J. et al. IRF5 is elevated in childhood-onset SLE and regulated by histone acetyltransferase and histone deacetylase inhibitors. Oncotarget 8, 47184–47194 (2017).
Karouzakis, E., Gay, R. E., Michel, B. A., Gay, S. & Neidhart, M. DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 60, 3613–3622 (2009).
Absher, D. M. et al. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 9, e1003678 (2013).
Oelke, K. et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 50, 1850–1860 (2004).
Sunahori, K. et al. The catalytic subunit of protein phosphatase 2A (PP2Ac) promotes DNA hypomethylation by suppressing the phosphorylated mitogen-activated protein kinase/extracellular signal-regulated kinase (ERK) kinase (MEK)/phosphorylated ERK/DNMT1 protein pathway in T-cells from controls and systemic lupus erythematosus patients. JBC 288, 21936–21944 (2013).
Gorelik, G., Sawalha, A. H., Patel, D., Johnson, K. & Richardson, B. T cell PKCδ kinase inactivation induces lupus-like autoimmunity in mice. Clin. Immunol. 158, 193–203 (2015).
Lashine, Y. A., Salah, S., Aboelenein, H. R. & Abdelaziz, A. I. Correcting the expression of miRNA-155 represses PP2Ac and enhances the release of IL-2 in PBMCs of juvenile SLE patients. Lupus 24, 240–247 (2015).
Kimura, T., Egawa, S. & Uemura, H. Personalized peptide vaccines and their relation to other therapies in urological cancer. Nat. Rev. Urol. 14, 501–510 (2017).
Punt, C. J. A., Koopman, M. & Vermeulen, L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat. Rev. Clin. Oncol. 14, 235–246 (2016).
Trynka, G. et al. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 45, 124–130 (2012).
Bujold, D. et al. The international human epigenome consortium data portal. Cell Syst. 3, 496–499.e2 (2016).
Chung, S. A. et al. Differential genetic associations for systemic lupus erythematosus based on anti–dsDNA autoantibody production. PLoS Genet. 7, e1001323 (2011).
Glossop, J. R. et al. DNA methylation at diagnosis is associated with response to disease-modifying drugs in early rheumatoid arthritis. Epigenomics 9, 419–428 (2017).
Cribbs, A. P. et al. Methotrexate restores regulatory T cell function through demethylation of the FOXP3 upstream enhancer in patients with rheumatoid arthritis. Arthritis Rheumatol. 67, 1182–1192 (2015).
Hammaker, D. et al. LBH gene transcription regulation by the interplay of an enhancer risk allele and DNA methylation in rheumatoid arthritis. Arthritis Rheumatol. 68, 2637–2645 (2016).
Sohn, C. et al. Prolonged TNFα primes fibroblast-like synoviocytes in a gene-specific manner by altering chromatin. Arthritis Rheumatol. 67, 86–95 (2015).
Lee, A. et al. Tumor necrosis factor α induces sustained signaling and a prolonged and unremitting inflammatory response in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 65, 928–938 (2013).
Lin, J. et al. A novel p53/microRNA-22/Cyr61 axis in synovial cells regulates inflammation in rheumatoid arthritis. Arthritis Rheumatol. 66, 49–59 (2014).
Philippe, L. et al. miR-20a regulates ASK1 expression and TLR4-dependent cytokine release in rheumatoid fibroblast-like synoviocytes. Ann. Rheum. Dis. 72, 1071–1079 (2013).
Trenkmann, M. et al. Expression and function of EZH2 in synovial fibroblasts: epigenetic repression of the Wnt inhibitor SFRP1 in rheumatoid arthritis. Ann. Rheum. Dis. 70, 1482–1488 (2011).
Stanczyk, J. et al. Altered expression of microRNA-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 63, 373–381 (2011).
Rhead, B. et al. Rheumatoid arthritis naive T cells share hypermethylation sites with synoviocytes. Arthritis Rheumatol. 69, 550–559 (2017).
Alexander, T. et al. FOXP3+ Helios+ regulatory T cells are expanded in active systemic lupus erythematosus. Ann. Rheum. Dis. 72, 1549–1558 (2013).
Hedrich, C. M. et al. cAMP response element modulator controls IL-2 and IL-17A expression during CD4 lineage commitment and subset distribution in lupus. Proc. Natl Acad. Sci. USA 109, 16606–16611 (2012).
Ding, S. et al. Decreased microRNA-142-3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum. 64, 2953–2963 (2012).
Zhang, Z. et al. Interferon regulatory factor 1 marks activated genes and can induce target gene expression in systemic lupus erythematosus. Arthritis Rheumatol. 67, 785–796 (2015).
Smith, S. et al. MicroRNA-302d targets IRF9 to regulate the IFN-induced gene expression in SLE. J. Autoimmun. 79, 105–111 (2017).
Zhao, M. et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann. Rheum. Dis. 75, 1998–2006 (2016).
Altorok, N., Tsou, P. S., Coit, P., Khanna, D. & Sawalha, A. H. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann. Rheum. Dis. 74, 1612–1620 (2015).
Dees, C. et al. The Wnt antagonists DKK1 and SFRP1 are downregulated by promoter hypermethylation in systemic sclerosis. Ann. Rheum. Dis. 73, 1232–1239 (2014).
Iwamoto, N. et al. Downregulation of miR-193b in systemic sclerosis regulates the proliferative vasculopathy by urokinase-type plasminogen activator expression. Ann. Rheum. Dis. 75, 303–310 (2016).
Lian, X. et al. DNA demethylation of CD40L in CD4+ T cells from women with systemic sclerosis: a possible explanation for female susceptibility. Arthritis Rheum. 64, 2338–2345 (2012).
Charras, A. et al. Cell-specific epigenome-wide DNA methylation profile in long-term cultured minor salivary gland epithelial cells from patients with Sjögren's syndrome. Ann. Rheum. Dis. 76, 625–628 (2017).
Mavragani, C. P. et al. Expression of long interspersed nuclear element 1 retroelements and induction of type I interferon in patients with systemic autoimmune disease. Arthritis Rheumatol. 68, 2686–2696 (2016).
Konsta, O. D. et al. Defective DNA methylation in salivary gland epithelial acini from patients with Sjögren's syndrome is associated with SSB gene expression, anti-SSB/LA detection, and lymphocyte infiltration. J. Autoimmun. 68, 30–38 (2016).
Alevizos, I., Alexander, S., Turner, R. J. & Illei, G. G. MicroRNA expression profiles as biomarkers of minor salivary gland inflammation and dysfunction in Sjögren's syndrome. Arthritis Rheum. 63, 535–544 (2011).
Altorok, N. et al. Genome-wide DNA methylation patterns in naive CD4+ T cells from patients with primary Sjögren's syndrome. Arthritis Rheumatol. 66, 731–739 (2014).
Imgenberg-Kreuz, J. et al. Genome-wide DNA methylation analysis in multiple tissues in primary Sjögren's syndrome reveals regulatory effects at interferon-induced genes. Ann. Rheum. Dis. 75, 2029–2036 (2016).
Miceli-Richard, C. et al. Overlap between differentially methylated DNA regions in blood B lymphocytes and genetic at-risk loci in primary Sjögren's syndrome. Ann. Rheum. Dis. 75, 933–940 (2016).
Latsoudis, H. et al. Differential expression of miR-4520a associated with pyrin mutations in familial Mediterranean fever (FMF). J. Cell. Physiol. 232, 1326–1336 (2017).
Mishra, N., Reilly, C. M., Brown, D. R., Ruiz, P. & Gilkeson, G. S. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J. Clin. Invest. 111, 539–552 (2003).
Hemmatazad, H. et al. Histone deacetylase 7, a potential target for the antifibrotic treatment of systemic sclerosis. Arthritis Rheum. 60, 1519–1529 (2009).
de Andres, M. C. et al. Assessment of global DNA methylation in peripheral blood cell subpopulations of early rheumatoid arthritis before and after methotrexate. Arthritis Res. Ther. 17, 233 (2015).
Garaud, S. et al. IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J. Immunol. 182, 5623–5632 (2009).
Plant, D. et al. Differential methylation as a biomarker of response to etanercept in patients with rheumatoid arthritis. Arthritis Rheumatol. 68, 1353–1360 (2016).
Author information
Authors and Affiliations
Contributions
Both authors researched data for the article, made substantial contributions to discussions of the content, wrote the article and reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Ballestar, E., Li, T. New insights into the epigenetics of inflammatory rheumatic diseases. Nat Rev Rheumatol 13, 593–605 (2017). https://doi.org/10.1038/nrrheum.2017.147
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrrheum.2017.147
- Springer Nature Limited
This article is cited by
-
Potential role of camel, mare milk, and their products in inflammatory rheumatic diseases
Rheumatology International (2024)
-
Innate immune memory in inflammatory arthritis
Nature Reviews Rheumatology (2023)
-
Age-related mechanisms in the context of rheumatic disease
Nature Reviews Rheumatology (2022)
-
Epigenetic regulation of B cells and its role in autoimmune pathogenesis
Cellular & Molecular Immunology (2022)
-
BCL-6 and EZH2 cooperate to epigenetically repress anti-inflammatory miR-142-3p/5p in lupus CD4+T cells
Cellular & Molecular Immunology (2021)