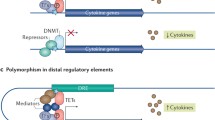
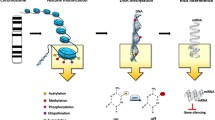
Abstract
Rheumatoid arthritis is a complex disorder that is characterized by irreversible and progressive destructions of joints, but its exact etiology remains mainly unknown. The occurrence and the progression of the disease entirely depend on environmental and genetic factors. In recent years, various epigenetic changes involving DNA methylation, histone modification, miRNA, X-chromosome inactivation, bromodomain, sirtuin, and many others were identified that were found to be linked to the activation and the aggressive phenotype in rheumatoid arthritis. Epigenetics is found to be one of the root causes, which brings changes in the heritable phenotype and is not determined by changes in the DNA sequences and understanding these epigenetic mechanisms and the pathogenesis of the disease can help in understanding the disease and various other possible ways for its control and/or prevention. The various epigenetic modification occurring are reversible and can be modulated by drugs, diet, and environmental factors. This article focuses on various epigenetic factors involved in the pathogenesis of rheumatoid arthritis. Further, various epigenetic therapies that might be successful in inhibiting these epigenetic modifications are summarized. Several therapeutic agents alter the epigenetic modifications occurring in various diseases and many of the epigenetic therapies are under pre-clinical and clinical trial. However, exploring these epigenetic prognostic biomarkers would give a broader perspective and provide more ideas and knowledge regarding the process and pathways through which the diseases occur, and also combining various therapeutic agents would show more beneficial and synergistic effects.


Similar content being viewed by others
Availability of data and material (data transparency)
Not Applicable (Review Article).
Code availability (software application or custom code)
Not Applicable.
Abbreviations
- AICDA:
-
Activation-induced cytidine deaminase
- BETi:
-
Bromodomain and extra-terminal
- CXCL12:
-
CXC motif chemokine ligand 12
- DNMT:
-
DNA methyltransferase
- FLS:
-
Fibroblast-like synoviocytes
- Foxp3:
-
Forkhead box p3
- HDAC:
-
Histone deacetylase
- IFNg:
-
Interferon gamma
- IL:
-
Interleukin
- IRAK1:
-
Interleukin q receptor-associated kinase 1
- IκBs:
-
Inhibitors of kappa B
- KDM6B:
-
Lysine (K)-specific demethylase 6B
- miR:
-
Mirco RNA
- NFκB:
-
Nuclear factor-kappa B
- OASF:
-
Osteoarthritis synovial fibroblast
- PBMC:
-
Peripheral blood mononuclear cells
- PI3K:
-
Phosphoinositide 3-kinase pathway
- RA:
-
Rheumatoid arthritis
- RASF:
-
Rheumatoid arthritis synovial fibroblast
- SF:
-
Synovial fibroblast
- SF:
-
Synovial fibroblast
- TF:
-
Transcription factors
- TGFb:
-
Transforming growth factor beta
- TLR2:
-
Toll like receptor 2
References
Karami J, Masoumi M, Khorramdelazad H, Bashiri H, Darvishi P, Sereshki HA et al (2020) Role of autophagy in the pathogenesis of rheumatoid arthritis: latest evidence and therapeutic approaches. Life Sci 254:117734. https://doi.org/10.1016/j.lfs.2020.117734
Edilova MI, Akram A, Abdul-Sater AA (2020) Innate immunity drives pathogenesis of rheumatoid arthritis. Biomed J. https://doi.org/10.1016/j.bj.2020.06.010
Intriago M, Maldonado G, Cárdenas J, Ríos C (2019) Clinical characteristics in patients with rheumatoid arthritis: differences between genders. Sci World J. https://doi.org/10.1155/2019/8103812
Nemtsova MV, Zaletaev DV, Bure IV, Mikhaylenko DS, Kuznetsova EB, Alekseeva EA et al (2019) Epigenetic changes in the pathogenesis of rheumatoid arthritis. Front Genet 10:1–13. https://doi.org/10.3389/fgene.2019.00570
Yap H-Y, Tee S, Wong M, Chow S-K, Peh S-C, Teow S-Y (2018) Pathogenic role of immune cells in rheumatoid arthritis: implications in clinical treatment and biomarker development. Cells 7:161. https://doi.org/10.3390/cells7100161
Cavalli G, Heard E (2019) Advances in epigenetics link genetics to the environment and disease. Nature 571:489–499. https://doi.org/10.1038/s41586-019-1411-0
Bustamante MF, Garcia-Carbonell R, Whisenant KD, Guma M (2017) Fibroblast-like synoviocyte metabolism in the pathogenesis of rheumatoid arthritis. Arthritis Res Ther 19:1–12. https://doi.org/10.1186/s13075-017-1303-3
Karami J, Aslani S, Tahmasebi MN, Mousavi MJ, Sharafat Vaziri A, Jamshidi A et al (2020) Epigenetics in rheumatoid arthritis; fibroblast-like synoviocytes as an emerging paradigm in the pathogenesis of the disease. Immunol Cell Biol 98:171–186. https://doi.org/10.1111/imcb.12311
Chang K, Yang SM, Kim SH, Han KH, Park SJ, Shin JI (2014) Smoking and rheumatoid arthritis. Int J Mol Sci 15:22279–22295. https://doi.org/10.3390/ijms151222279
Frank-Bertoncelj M, Klein K, Gay S (2017) Interplay between genetic and epigenetic mechanisms in rheumatoid arthritis. Epigenomics 9:493–504. https://doi.org/10.2217/epi-2016-0142
Ciechomska M, Roszkowski L, Maslinski W (2019) DNA methylation as a future therapeutic and diagnostic target in rheumatoid arthritis Cells 8(9):953. https://doi.org/10.3390/cells8090953
Glant TT, Mikecz K, Rauch TA (2014) Epigenetics in the pathogenesis of rheumatoid arthritis. BMC Med 12:1–5. https://doi.org/10.1186/1741-7015-12-35
Ziegler SF, Buckner JH (2009) FOXP3 and the regulation of Treg/Th17 differentiation. Microbes Infect 11:594–598. https://doi.org/10.1016/j.micinf.2009.04.002
Nygaard G, Firestein GS (2020) Restoring synovial homeostasis in rheumatoid arthritis by targeting fibroblast-like synoviocytes. Nat Rev Rheumatol 16:316–333. https://doi.org/10.1038/s41584-020-0413-5
Guo S, Xu L, Chang C, Zhang R, Jin Y, He D (2020) Epigenetic regulation mediated by methylation in the pathogenesis and precision medicine of rheumatoid arthritis. Front Genet 11:1–9. https://doi.org/10.3389/fgene.2020.00811
Ahmadi M, Gharibi T, Dolati S, Rostamzadeh D, Aslani S, Baradaran B et al (2017) Epigenetic modifications and epigenetic based medication implementations of autoimmune diseases. Biomed Pharmacother 87:596–608. https://doi.org/10.1016/j.biopha.2016.12.072
Papin C, Ibrahim A, Le Gras S, Velt A, Stoll I, Jost B et al (2017) Combinatorial DNA methylation codes at repetitive elements. Genome Res 27:934–946. https://doi.org/10.1101/gr.213983.116
O’Neill RJ, Vrana PB, Rosenfeld CS (2014) Maternal methyl supplemented diets and effects on offspring health. Front Genet 5:1–11. https://doi.org/10.3389/fgene.2014.00289
Rider CF, Carlsten C (2019) Air pollution and DNA methylation: effects of exposure in humans. Clin Epigenet 11:1–15. https://doi.org/10.1186/s13148-019-0713-2
Karagianni P, Tzioufas AG (2019) Epigenetic perspectives on systemic autoimmune disease. J Autoimmun. https://doi.org/10.1016/j.jaut.2019.102315
Gujar H, Weisenberger DJ, Liang G (2019) The roles of human DNA methyltransferases and their isoforms in shaping the epigenome. Genes (Basel). https://doi.org/10.3390/genes10020172
Brandt B, Rashidiani S, Bán Á, Rauch TA (2019) DNA methylation-governed gene expression in autoimmune arthritis. Int J Mol Sci. https://doi.org/10.3390/ijms20225646
Zhu H, Wu LF, Mo XB, Lu X, Tang H, Zhu XW et al (2019) Rheumatoid arthritis-associated DNA methylation sites in peripheral blood mononuclear cells. Ann Rheum Dis 78:36–42. https://doi.org/10.1136/annrheumdis-2018-213970
Karouzakis E, Gay RE, Gay S, Neidhart M (2012) Increased recycling of polyamines is associated with global DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum 64:1809–1817. https://doi.org/10.1002/art.34340
Karouzakis E, Raza K, Kolling C, Buckley CD, Gay S, Filer A et al (2018) Analysis of early changes in DNA methylation in synovial fibroblasts of RA patients before diagnosis. Sci Rep 8:1–6. https://doi.org/10.1038/s41598-018-24240-2
Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A et al (2012) In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. https://doi.org/10.1371/journal.pgen.1002750
Glossop JR, Emes RD, Nixon NB, Haworth KE, Packham JC, Dawes PT et al (2014) Genome-wide DNA methylation profiling in rheumatoid arthritis identifies disease-associated methylation changes that are distinct to individual T- and B-lymphocyte populations. Epigenetics 9:1228–1237. https://doi.org/10.4161/epi.29718
Liu CC, Fang TJ, Ou TT, Wu CC, Li RN, Lin YC et al (2011) Global DNA methylation, DNMT1, and MBD2 in patients with rheumatoid arthritis. Immunol Lett 135:96–99. https://doi.org/10.1016/j.imlet.2010.10.003
Kennedy A, Schmidt EM, Cribbs AP, Penn H, Amjadi P, Syed K et al (2014) A novel upstream enhancer of FOXP3, sensitive to methylation-induced silencing, exhibits dysregulated methylation in rheumatoid arthritis Treg cells. Eur J Immunol 44:2968–2978. https://doi.org/10.1002/eji.201444453
Zhao M, Wang Z, Yung S, Lu Q (2015) Epigenetic dynamics in immunity and autoimmunity. Int J Biochem Cell Biol. https://doi.org/10.1016/j.biocel.2015.05.022
Angiolilli C, Kabala PA, Grabiec AM, Van Baarsen IM, Ferguson BS, García S et al (2017) Histone deacetylase 3 regulates the inflammatory gene expression programme of rheumatoid arthritis fibroblast-like synoviocytes. Ann Rheum Dis 76:277–285. https://doi.org/10.1136/annrheumdis-2015-209064
Karouzakis E, Trenkmann M, Gay RE, Michel BA, Gay S, Neidhart M (2014) Epigenome analysis reveals TBX5 as a novel transcription factor involved in the activation of rheumatoid arthritis synovial fibroblasts. J Immunol 193:4945–4951. https://doi.org/10.4049/jimmunol.1400066
Picascia A, Grimaldi V, Pignalosa O, De Pascale MR, Schiano C, Napoli C (2015) Epigenetic control of autoimmune diseases: from bench to bedside. Clin Immunol 157:1–15. https://doi.org/10.1016/j.clim.2014.12.013
Miao CG, Yang YY, He X, Li J (2013) New advances of DNA methylation and histone modifications in rheumatoid arthritis, with special emphasis on MeCP2. Cell Signal 25:875–882. https://doi.org/10.1016/j.cellsig.2012.12.017
Trenkmann M, Brock M, Gay RE, Kolling C, Speich R, Michel BA et al (2011) Expression and function of EZH2 in synovial fibroblasts: epigenetic repression of the Wnt inhibitor SFRP1 in rheumatoid arthritis. Ann Rheum Dis 70:1482–1488. https://doi.org/10.1136/ard.2010.143040
Webster AP, Plant D, Ecker S, Zufferey F, Bell JT, Feber A et al (2018) Increased DNA methylation variability in rheumatoid arthritis-discordant monozygotic twins. Genome Med 10:1–12. https://doi.org/10.1186/s13073-018-0575-9
Wang Y, He J, Liao M, Hu M, Li W, Ouyang H et al (2019) An overview of Sirtuins as potential therapeutic target: structure, function and modulators. Eur J Med Chem 161:48–77. https://doi.org/10.1016/j.ejmech.2018.10.028
Kong S, Yeung P, Fang D (2013) The class III histone deacetylase sirtuin 1 in immune suppression and its therapeutic potential in rheumatoid arthritis. J Genet Genomics 40:347–354. https://doi.org/10.1016/j.jgg.2013.04.001
Moon MH, Jeong JK, Lee YJ, Seol JW, Jackson CJ, Park SY (2013) SIRT1, a class III histone deacetylase, regulates TNF-α-induced inflammation in human chondrocytes. Osteoarthr Cartil 21:470–480. https://doi.org/10.1016/j.joca.2012.11.017
Mendes KL, Lelis DDF, Santos SHS (2017) Nuclear sirtuins and inflammatory signaling pathways. Cytokine Growth Factor Rev 38:98–105. https://doi.org/10.1016/j.cytogfr.2017.11.001
Niederer F, Ospelt C, Brentano F, Hottiger MO, Gay RE, Gay S et al (2011) SIRT1 overexpression in the rheumatoid arthritis synovium contributes to proinflammatory cytokine production and apoptosis resistance. Ann Rheum Dis 70:1866–1873. https://doi.org/10.1136/ard.2010.148957
Kulikowski E, Rakai BD, Wong NCW (2021) Inhibitors of bromodomain and extra-terminal proteins for treating multiple human diseases. Med Res Rev 41:223–245. https://doi.org/10.1002/med.21730
Xiao Y, Liang L, Huang M, Qiu Q, Zeng S, Shi M et al (2016) Bromodomain and extra-terminal domain bromodomain inhibition prevents synovial inflammation via blocking IκB kinase-dependent NF-κB activation in rheumatoid fibroblast-like synoviocytes. Rheumatol (UK) 55:173–184. https://doi.org/10.1093/rheumatology/kev312
Klein K (2018) Bromodomain protein inhibition: a novel therapeutic strategy in rheumatic diseases. RMD Open 4:e000744. https://doi.org/10.1136/rmdopen-2018-000744
Martin GV, Kanaan SB, Hemon MF, Azzouz DF, El Haddad M, Balandraud N et al (2019) Mosaicism of XX and XXY cells accounts for high copy number of toll like receptor 7 and 8 genes in peripheral blood of men with rheumatoid arthritis. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-49309-4
Ballestar E (2011) Epigenetic alterations in autoimmune rheumatic diseases. Nat Rev Rheumatol 7:263–271. https://doi.org/10.1038/nrrheum.2011.16
Takheaw N, Earwong P, Laopajon W, Pata S, Kasinrerk W (2019) Interaction of CD99 and its ligand upregulates IL-6 and TNF-α upon T cell activation. PLoS ONE 14:1–19. https://doi.org/10.1371/journal.pone.0217393
Moran-Moguel MC, Del RSP, Mayorquin-Galvan EE, Zavala-Cerna MG (2018) Rheumatoid arthritis and miRNAs: a critical review through a functional view. J Immunol Res. https://doi.org/10.1155/2018/2474529
Corsiero E, Marrelli A (2018) An update on research advances in rheumatoid arthritis: from clinic to basic science. J Lab Precis Med 3:54–54. https://doi.org/10.21037/jlpm.2018.06.03
Dong L, Wang X, Tan J, Li H, Qian W, Chen J et al (2014) Decreased expression of microRNA-21 correlates with the imbalance of Th17 and Treg cells in patients with rheumatoid arthritis. J Cell Mol Med 18:2213–2224. https://doi.org/10.1111/jcmm.12353
O’Brien J, Hayder H, Zayed Y, Peng C (2018) Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 9:1–12. https://doi.org/10.3389/fendo.2018.00402
Ayeldeen G, Nassar Y, Ahmed H, Shaker O, Gheita T (2018) Possible use of miRNAs-146a and -499 expression and their polymorphisms as diagnostic markers for rheumatoid arthritis. Mol Cell Biochem 449:145–156. https://doi.org/10.1007/s11010-018-3351-7
Fu H, Hu D, Zhang L, Tang P (2018) Role of extracellular vesicles in rheumatoid arthritis. Mol Immunol 93:125–132. https://doi.org/10.1016/j.molimm.2017.11.016
Lu MC, Yu CL, Chen HC, Yu HC, Huang HB, Lai NS (2014) Increased miR-223 expression in T cells from patients with rheumatoid arthritis leads to decreased insulin-like growth factor-1-mediated interleukin-10 production. Clin Exp Immunol 177:641–651. https://doi.org/10.1111/cei.12374
Murata K, Yoshitomi H, Furu M, Ishikawa M, Shibuya H, Ito H et al (2014) Microrna-451 down-regulates neutrophil chemotaxis via p38 mapk. Arthritis Rheumatol 66:549–559. https://doi.org/10.1002/art.38269
Sánchez-Pernaute O (2010) Epigenetic therapies, a step beyond biologics for rheumatoid arthritis. Reumatol Clínica (Engl Ed) 6:306–310. https://doi.org/10.1016/s2173-5743(10)70072-1
Krishna V, Yin X, Song Q, Walsh A, Pocalyko D, Bachman K et al (2021) Integration of the transcriptome and genome-wide landscape of BRD2 and BRD4 binding motifs identifies key superenhancer genes and reveals the mechanism of bet inhibitor action in rheumatoid arthritis synovial fibroblasts. J Immunol 206:422–431. https://doi.org/10.4049/jimmunol.2000286
Kantarjian HM, Roboz GJ, Kropf PL, Yee KWL, O’Connell CL, Tibes R et al (2017) Guadecitabine (SGI-110) in treatment-naive patients with acute myeloid leukaemia: phase 2 results from a multicentre, randomised, phase 1/2 trial. Lancet Oncol 18:1317–1326. https://doi.org/10.1016/S1470-2045(17)30576-4
Marques-Magalhães Â, Graça I, Henrique R, Jerónimo C (2018) Targeting DNA methyltranferases in urological tumors. Front Pharmacol. https://doi.org/10.3389/fphar.2018.00366
Plummer R, Vidal L, Griffin M, Lesley M, De Bono J, Coulthard S et al (2009) Phase I study of MG98, an oligonucleotide antisense inhibitor of human DNA methyltransferase 1, given as a 7-day infusion in patients with advanced solid tumors. Clin Cancer Res 15:3177–3183. https://doi.org/10.1158/1078-0432.CCR-08-2859
Lin RK, Hsu CH, Wang YC (2007) Mithramycin A inhibits DNA methyltransferase and metastasis potential of lung cancer cells. Anticancer Drugs 18:1157–1164. https://doi.org/10.1097/CAD.0b013e3282a215e9
Chen L, Jin T, Zhu K, Piao Y, Quan T, Quan C et al (2017) PI3K/mTOR dual inhibitor BEZ235 and histone deacetylase inhibitor Trichostatin A synergistically exert anti-tumor activity in breast cancer. Oncotarget 8:11937–11949. https://doi.org/10.18632/oncotarget.14442
Kirschbaum MH, Foon KA, Frankel P, Ruel C, Pulone B, Tuscano JM et al (2014) A phase 2 study of belinostat (PXD101) in patients with relapsed or refractory acute myeloid leukemia or patients over the age of 60 with newly diagnosed acute myeloid leukemia: a California Cancer Consortium Study. Leuk Lymphoma 55:2301–2304. https://doi.org/10.3109/10428194.2013.877134
Sivaraj D, Green MM, Gasparetto C (2017) Panobinostat for the management of multiple myeloma. Futur Oncol 13:477–488. https://doi.org/10.2217/fon-2016-0329
Klein K, Gay S (2013) Epigenetic modifications in rheumatoid arthritis, a review. Curr Opin Pharmacol. https://doi.org/10.1016/j.coph.2013.01.007
Mandl-Weber S, Meinel FG, Jankowsky R, Oduncu F, Schmidmaier R, Baumann P (2010) The novel inhibitor of histone deacetylase resminostat (RAS2410) inhibits proliferation and induces apoptosis in multiple myeloma (MM) cells. Br J Haematol 149:518–528. https://doi.org/10.1111/j.1365-2141.2010.08124.x
He B, Dai L, Zhang X, Chen D, Wu J, Feng X et al (2018) The HDAC inhibitor quisinostat (JNJ-26481585) supresses hepatocellular carcinoma alone and synergistically in combination with sorafenib by G0/G1 phase arrest and apoptosis induction. Int J Biol Sci 14:1845–1858. https://doi.org/10.7150/ijbs.27661
Deutsch E, Moyal ECJ, Gregorc V, Zucali PA, Menard J, Soria JC et al (2017) A phase 1 dose-escalation study of the oral histone deacetylase inhibitor abexinostat in combination with standard hypofractionated radiotherapy in advanced solid tumors. Oncotarget 8:56199–56209. https://doi.org/10.18632/oncotarget.14147
Cao J, Lv W, Wang L, Xu J, Yuan P, Huang S et al (2018) Ricolinostat (ACY-1215) suppresses proliferation and promotes apoptosis in esophageal squamous cell carcinoma via miR-30d/PI3K/AKT/mTOR and ERK pathways. Cell Death Dis. https://doi.org/10.1038/s41419-018-0788-2
Eigl BJ, North S, Winquist E, Finch D, Wood L, Sridhar SS et al (2015) A phase II study of the HDAC inhibitor SB939 in patients with castration resistant prostate cancer: NCIC clinical trials group study IND195. Invest New Drugs 33:969–976. https://doi.org/10.1007/s10637-015-0252-4
De Bono JS, Kristeleit R, Tolcher A, Fong P, Pacey S, Karavasilis V et al (2008) Phase I pharmacokinetic and pharmacodynamic study of LAQ824, a hydroxamate histone deacetylase inhibitor with a heat shock protein-90 inhibitory profile, in patients with advanced solid tumors. Clin Cancer Res 14:6663–6673. https://doi.org/10.1158/1078-0432.CCR-08-0376
Furumai R, Matsuyama A, Kobashi N, Lee KH, Nishiyama M, Nakajima H et al (2002) FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res 62:4916–4921
Loprevite M, Tiseo M, Grossi F, Scolaro T, Semino C, Pandolfi A et al (2005) In vitro study of CI-994, a histone deacetylase inhibitor, in non-small cell lung cancer cell lines. Oncol Res 15:39–48. https://doi.org/10.3727/096504005775082066
Chan E, Chiorean EG, O’Dwyer PJ, Gabrail NY, Alcindor T, Potvin D et al (2018) Phase I/II study of mocetinostat in combination with gemcitabine for patients with advanced pancreatic cancer and other advanced solid tumors. Cancer Chemother Pharmacol 81:355–364. https://doi.org/10.1007/s00280-017-3494-3
Zhijun H, Shusheng W, Han M, Jianping L, Li-sen Q, Dechun L (2016) Pre-clinical characterization of 4SC-202, a novel class I HDAC inhibitor, against colorectal cancer cells. Tumor Biol 37:10257–10267. https://doi.org/10.1007/s13277-016-4868-6
Mawatari T, Ninomiya I, Inokuchi M, Harada S, Hayashi H, Oyama K et al (2015) Valproic acid inhibits proliferation of HER2-expressing breast cancer cells by inducing cell cycle arrest and apoptosis through Hsp70 acetylation. Int J Oncol 47:2073–2081. https://doi.org/10.3892/ijo.2015.3213
Shi X, Zheng C, Li C, Hou K, Wang X, Yang Z et al (2018) 4-Phenybutyric acid promotes gastric cancer cell migration via histone deacetylase inhibition-mediated HER3/HER4 up-regulation. Cell Biol Int 42:53–62. https://doi.org/10.1002/cbin.10866
Vojinovic J, Damjanov N, D’Urzo C, Furlan A, Susic G, Pasic S et al (2011) Safety and efficacy of an oral histone deacetylase inhibitor in systemic-onset juvenile idiopathic arthritis. Arthritis Rheum 63:1452–1458. https://doi.org/10.1002/art.30238
Lin H-S, Hu C-Y, Chan H-Y, Liew Y-Y, Huang H-P, Lepescheux L et al (2007) Anti-rheumatic activities of histone deacetylase (HDAC) inhibitors in vivo in collagen-induced arthritis in rodents. Br J Pharmacol 150:862–872. https://doi.org/10.1038/sj.bjp.0707165
Miao C, Yang Y, He X, Li J (2013) New advances of DNA methylation and histone modifications in rheumatoid arthritis, with special emphasis on MeCP2. Cell Signal 25:875–882. https://doi.org/10.1016/j.cellsig.2012.12.017
Chen YJ, Wang WH, Wu WY, Hsu CC, Wei LR, Wang SF et al (2017) Novel histone deacetylase inhibitor AR-42 exhibits antitumor activity in pancreatic cancer cells by affecting multiple biochemical pathways. PLoS ONE 12:1–18. https://doi.org/10.1371/journal.pone.0183368
Van Der Ree MH, Van Der Meer AJ, Van Nuenen AC, De Bruijne J, Ottosen S, Janssen HL et al (2016) Miravirsen dosing in chronic hepatitis C patients results in decreased microRNA-122 levels without affecting other microRNAs in plasma. Aliment Pharmacol Ther 43:102–113. https://doi.org/10.1111/apt.13432
van der Ree MH, de Vree JM, Stelma F, Willemse S, van der Valk M, Rietdijk S et al (2017) Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: a phase 1B, double-blind, randomised controlled trial. Lancet 389:709–717. https://doi.org/10.1016/S0140-6736(16)31715-9
Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J et al (2017) Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs 35:180–188. https://doi.org/10.1007/s10637-016-0407-y
Seto AG, Beatty X, Lynch JM, Hermreck M, Tetzlaff M, Duvic M et al (2018) Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br J Haematol 183:428–444. https://doi.org/10.1111/bjh.15547
Cheng Y, He C, Wang M, Ma X, Mo F, Yang S et al (2019) Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct Target Ther. https://doi.org/10.1038/s41392-019-0095-0
Bondarev AD, Attwood MM, Jonsson J, Chubarev VN, Tarasov VV, Schiöth HB (2021) Recent developments of HDAC inhibitors: emerging indications and novel molecules. Br J Clin Pharmacol. https://doi.org/10.1111/bcp.14889
Zhang S, Cheng Z, Wang Y, Han T (2021) The Risks of miRNA therapeutics: in a drug target perspective. Drug Des Dev Ther 15:721–733. https://doi.org/10.2147/DDDT.S288859
Shorstova T, Foulkes WD, Witcher M (2021) Achieving clinical success with BET inhibitors as anti-cancer agents. Br J Cancer 124:1478–1490. https://doi.org/10.1038/s41416-021-01321-0
Funding
None.
Author information
Authors and Affiliations
Contributions
Conceptualization: [LKB]; Writing—original draft preparation: [RRB]; Writing—review and editing: [LKB].
Corresponding author
Ethics declarations
Conflict of interest
None.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
No part of the final manuscript, including ideas, tables and figures, is copied or published elsewhere in whole or in part.
Rights and permissions
About this article
Cite this article
Barik, R.R., Bhatt, L.K. Emerging epigenetic targets in rheumatoid arthritis. Rheumatol Int 41, 2047–2067 (2021). https://doi.org/10.1007/s00296-021-04951-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00296-021-04951-y