Abstract
MicroRNAs (miRNAs) have important roles in the regulation of genes; however, for trees few studies have explored the potential impact of the interactions between miRNAs and their target genes. Here, we performed transcript profiling and association genetics (single-SNP, haplotype-based and multi-SNP associations) to study the genetic regulatory relationship of Pto-miR257 and its 12 target genes in 435 individuals of a natural population of Populus tomentosa. Expression profiling of Pto-miR257 and its targets showed a negative relationship between their expression levels. Of the 61 single-nucleotide polymorphisms (SNPs) detected in Pto-miR257, 6 in the pre-mature region strongly affected its secondary stability and 1 in the mature region could alter its target spectrum. Among the 1029 SNPs in the targets, 3 were located in target sites that could change the binding affinity of Pto-miR257. Single-SNP association analysis revealed that SNPs in Pto-miR257 and target genes associated with both growth and wood property traits, in agreement with haplotype-based identifications. Multi-SNP association found that 10 targets shared at least one common trait with Pto-miR257, with phenotypic variance from 0.5 to 8.5%, suggesting a possible internal genetic interaction between them. Epistasis analysis showed significant epistatic interactions among Pto-miR257 and its targets. Therefore, our study demonstrated Pto-miR257 and its 12 targets had roles in wood formation and revealed the genetic interaction network between the miRNA and its targets under additive, dominant and epistatic models. Thus, association genetics can be used to decipher the interactions between miRNAs and their target genes and to help understand the genetic architecture of complex traits.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs), the endogenous ∼22-nucleotide noncoding RNAs, can affect gene expression by targeting mRNAs for cleavage or translational repression (Rogers and Chen, 2013). Previous studies have verified the important regulatory roles of miRNAs in biology (Kwon et al., 2013; Rogers and Chen, 2013; Sun and Lai, 2013). Experimental and computational approaches, especially in herbaceous plants, have illuminated the diverse functions of miRNAs in plant development (Rogers and Chen, 2013). However, few studies have explored the potential impact of miRNAs during the growth of long-lived perennial plants (Robischon et al., 2011; Lu et al., 2013). Recently, several studies have used reverse genetics on woody plants to reveal the biological importance of miRNA–target interactions in tree growth and wood formation (Robischon et al., 2011; Lu et al., 2013).
Because genetic modification remains difficult in trees, elucidating the contributions of miRNA-mediated gene regulation to phenotypic variation will require studies to use methods beyond transgenic approaches. Moreover, because one miRNA can target multiple genes, the regulatory interactions of miRNAs form a complex network that includes miRNA–target and target–target effects. For instance, a study in humans indicated that miR-137 and its schizophrenia-related targets (CSMD1, C10orf26, CACNA1C, and TCF4) would function together as a regulatory network (Kwon et al., 2013), and this seems impossible to elucidate by simple application of reverse-genetic methods. Thus, forest trees may require a novel strategy for examining the role of miRNA-target interactions underlying quantitative traits.
Single-nucleotide polymorphisms (SNPs) commonly occurred in miRNA genes, including the flanking regions, the precursor regions (pre-miRNA) and the mature regions of miRNA genes in human (Yang et al., 2014), animal (Lei et al., 2011) and plant genomes (Liu et al., 2013). SNPs also occur in the miRNA binding sites of target genes (Dusl et al., 2015). SNPs located in pre-mature miRNA regions likely cause complex effects by altering miRNA processing and target interactions (Yang et al., 2014). SNPs in mature miRNAs or their binding sites in target genes could also change the miRNA–target binding affinities, thus affecting the target spectrum of miRNAs. For example, studies on human miRNA SNPs showed that patients with congenital myasthenic syndrome had a 3′ untranslated region mutation in the GFPT1 gene that could lead to illegitimate binding of an miRNA, thus resulting in reduced protein expression (Dusl et al., 2015). Except the pre-mature region, SNPs within the flanking sequence of miRNA genes or the sequences around target sites might also affect target recognition (Lei et al., 2011). These SNPs represent genetic variation that can modulate the regulatory interaction between miRNAs and their target genes and probably affect phenotypic variation. However, at present, few SNPs within miRNA genes and miRNA targets genes have been investigated in plants, particularly in trees (Yang et al., 2015).
Long-term natural selection in tree species produced abundant allelic variations. Detection of complicated variants of miRNAs and their target genes will help to explore the roles miRNAs play in regulating their target mRNAs and contribute to identification of functional mutations. Linkage disequilibrium (LD)-based association mapping, particularly candidate gene-based SNP association, has been used to examine associations between natural allelic variation and phenotype in tree species such as conifers and eucalyptus (Thumma et al., 2005; Beaulieu et al., 2011). However, previous studies always only considered the genotypic effects of single markers, and rarely took into account the combined effects of three genetic interactions (additive, dominant and epistatic) that collectively contribute to trait diversity, especially for prediction of allelic epistatic effects among genes that could inform our understanding of their genetic interactions.
In the present study, we identified Pto-miR257, a new miRNA exhibiting high expression levels in mature xylem of Populus tomentosa. By target gene prediction and transcriptome sequencing, we found 12 of its targets showed high and differential expression in different part of the stem vascular tissues of P. tomentosa. Expression patterns of Pto-miR257 and the 12 targets in stem indicated that they may function in wood formation. Here, we investigated the nature of genetic variance for Pto-miR257 and its 12 targets with 10 quantitative traits, using single- and multi-SNP association, together with haplotype-based association approaches, in a natural population of P. tomentosa. We identified SNPs that significantly associated with growth and wood properties; some also strongly affect the secondary stability of Pto-miR257 transcripts and change target accessibility. Moreover, tissue-specific analysis of transcript abundances of Pto-miR257 and its targets provide a better understanding of the complex miRNA–mRNA and mRNA–mRNA interaction network. Thus, our study provided a viable approach for exploring the genetic architecture of complex traits involving miRNA–target interactions that can be used for miRNAs studies in humans, animals and plants, including trees.
Materials and methods
Population and phenotypic data
A clonal arboretum of P. tomentosa was established in Guan Xian County, Shandong Province, China (36°23′N, 115°47′E), containing 1047 unrelated individuals collected from the entire nature distribution region (∼1 million km2) of P. tomentosa in 1982. Ten quantitative traits were measured in 435 individuals randomly selected from the clonal arboretum (Supplementary Data S1): the tree growth traits were diameter at breast height (DBH), tree height (H) and stem volume (V); the wood chemical traits were α-cellulose content, holocellulose content, hemicellulose content and lignin content; and wood physical traits were fiber length (FL), fiber width (FW) and microfiber angle (MFA). Data for tree H and D at breast height were collected during field surveys in 2011. FL and FW were measured using a Color CCTV Camera (Panasonic SDII, Osaka, Japan), and MFA was measured using an Xray powder diffractometer (Philips, Eindhoven, The Netherlands). Four reference standard procedures (GB/T2677.8–1994, GB/T2677.10–1995, GB/T 744–2004 and FZ/T50010.4–1998) were consulted to test for contents of wood chemical traits. Details of the measurement of these phenotypic data have been described by Du et al. (2014).
Identification and isolation of Pto-miR257 and its target genes
Identification and isolation of Pto-miR257
As biological replicates, three 30-year-old clones from one genotype of P. tomentosa were selected in this study. Total RNA was extracted from the stem cambium, developing xylem and mature xylem of each individual, using the Plant Qiagen RNeasy kit (Qiagen China, Shanghai, China) as described in the manufacturer’s instructions. Extracted RNA was used for construction of small RNA libraries and small RNAs were sequenced using an Illumina HiSeq 2000 (Illumina, San Diego, CA, USA) at the Shanghai Bio Institute (Shanghai, China). We used the prediction software Mireap (https://sourceforge.net/projects/mireap/) to predict novel miRNAs by exploring the secondary structures, DL1 cleavage sites and minimum free energies of the unannotated small RNA tags that could be mapped to the Populus genome. Basic criteria (Meyers et al., 2008) were used for selecting the potential novel miRNAs. Secondary structure of the Pto-miR257 precursor sequence was assessed by RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi).
For the candidate gene, Pto-miR257, the sequencing results yielded the 23-nucleotide mature sequence. By BLAST (Basic Local Alignment Search Tool) against the P. trichocarpa genome sequence, we obtained the Ptr-miR257 gene, including 167-bp sequence of the pre-mature miRNA and 400 bp of flanking sequence on each side. PCR amplification was carried out to isolate the Pto-miR257 gene in P. tomentosa. Total genomic DNA was extracted from fresh young leaves of 40 individuals described above and primer pairs were designed based on the 967-bp genome sequence from the P. trichocarpa genome. PCR was performed in a reaction volume of 25 μl containing 20 ng genomic DNA, 0.8 U Taq DNA polymerase (Promega, Beijing, China), 50 ng forward primer, 50 ng reverse primer, 1 × PCR buffer (Promega) and 0.2 mM dNTPs (Promega) with the following conditions: 95 °C hot start for 5 min; 95 °C denaturation for 30 s, 56 °C annealing for 30 s and 72 °C extension for 1 min for 30 cycles; and a final extension of 5 min at 72 °C. The purified PCR products were then ligated into the pGEMH-T Easy Vector (Promega) and sequenced using the Big Dye Terminator version 3.1 Cycle Sequencing kit (Applied Biosystems, Beijing, China). The genomic DNA sequence of the Pto-miR257 gene was submitted to GenBank and the accession number is shown in Supplementary Table S1.
Identification and isolation of the target genes of Pto-miR257
For high-throughput transcriptome sequencing of stem tissues (cambium, developing xylem and mature xylem), we used the extracted RNA described above to construct pooled samples. High-quality reads were mapped to the P. trichocarpa genome v2.0 and the expression of transcripts was calculated using FPKM (fragments per kilobase of transcript per million fragments) for normalized. Differentially expressed genes between the three tissues were determined according to fold change (⩾2) with P⩽0.05. Then, psRNATarget analysis (Dai and Zhao, 2011) (http://plantgrn.noble.org/psRNATarget/) was performed on a P. trichocarpa genomic library for the prediction of Pto-miR257 putative target genes via complementary base pairing interactions. Each of the 12 potential targets that have high or differential abundance in stem was cloned and sequenced by amplification from the genomic DNA of the 40 individuals of P. tomentosa, using the PCR method described in Zhang et al. (2010) (Supplementary Table S1 and Supplementary Data S2). All genomic DNA sequences were submitted to GenBank and the accession numbers are shown in Supplementary Table S1.
Degradome sequencing of three pooled stem tissues (cambium, developing xylem and mature xylem) from P. tomentosa was performed to examine the regulatory relationship between miRNAs and mRNAs. The extracted RNA described above was used to construct degradome libraries, which were sequenced by the Illumina HiSeq 2000 (Illumina). We used the fastx toolkit to exclude low-quality reads and adapter sequences. Then, the Cleaveland software (http://sites.psu.edu/axtell/software/cleaveland4/) was used to map degradome reads to the P. trichocarpa transcript database of JGI Phytozome V 7.0 (ftp://ftp.jgi-psf.org/pub/compgen/phytozome/v7.0/Ptrichocarpa) and the true miRNA cleavage site was distinguished from background noise with a target plot.
Measurement of transcript levels
Real-time quantitative PCR was performed on a 7500 Fast Real-Time PCR System (Bio-Rad, Hercules, CA, USA) using the SYBR Premix Ex Taq (TaKaRa, Dalian, China) as described in the manufacturer’s instructions. The amplification reactions were performed as described by Zhang et al. (2010) for target genes. For Pto-miR257, reactions were carried out according to Song et al. (2013). The melting curve was used to check the specificity of the amplified fragments and all data were analyzed using the Opticon Monitor Analysis software 3.1 tool (Bio-Rad), following the manual protocol. Primer Express 3.0 software (Applied Biosystems) was used to design the specific primer pairs for Pto-miR257 and its targets (Supplementary Table S2). Poplar Actin (Accession Number: EF145577) was used as the internal control for measurements of target gene expression. The relative level of Pto-miR257 was measured and standardized to 5.8S rRNA as described by Song et al. (2013). All reactions were conducted in triplicate technical and triplicate biological repetitions. Differential expression across genotypic classes was tested by analysis of variance. The complementary DNA templates for different tissues were obtained by reverse transcription of total RNA. For differential expression analysis, samples that were used for total RNA extraction were collected from different individuals. For tissue-specific expression analysis, total RNAs were extracted from nine tissues of 1-year-old P. tomentosa clone ‘LM50’: bark, root, shoot apex, cambium, developing xylem, mature xylem, phloem, mature leaf and young leaf. For each tissue, three individuals were used as biological replicates. For transcript analysis of SNP genotypes of the 44 significant markers that were identified by single-SNP-based associations, total RNAs were extracted from secondary xylem of 132 individuals (3 for each genotype) selected from the association population. To quantify target mRNA levels with different genotypic combinations of Pto-miR257-SNP56 and PtoDUF248-1-SNP33, 27 individuals (3 for each genotype pair) from the association population were selected for extracting total RNAs from the secondary xylem.
SNP identification and genotyping and LD analysis
To identify SNPs in Pto-miR257 and its 12 target genes, we aligned and compared the sequences from 40 unrelated individuals from the association population of P. tomentosa using the software MEGA 5.0 (Tamura et al. 2011), without considering insertions/deletions (INDELs). DnaSP, version 4.0 (Rozas et al. 2003), was used to identify SNPs and analyze the nucleotide polymorphisms. Nucleotide diversity was estimated by both the average number of segregating sites per site (θw) and the average number of pairwise differences per site between sequences (π). For genotyping, total genomic DNA was isolated from fresh young leaves of the 435 individuals from the association population. Subsequently, common SNPs (minor allele frequency >10%) were genotyped by single-nucleotide primer extension using a Beckman Coulter (Franklin Lakes, NJ, USA) sequencing system following the manufacturer’s instructions. To compare whether the SNP could affect the accessibility of the mRNA target site to miRNA, target accessibility was calculated by psRNATarget (Dai and Zhao, 2011). The r2 value between pairs of common SNPs (frequencies >10%) in the candidate gene was calculated by using TASSEL Ver. 2.0.1 (http://www.maizegenetics.net/) and the decay of LD with physical distance (base pairs) between informative SNPs within each gene was estimated by nonlinear regression analysis (Hill and Robertson, 1968). LD among the common SNPs (frequencies >10%) within the 13 genes was evaluated using genotype data from 435 individuals in the association population by the software package HAPLOVIEW (http://www.broad.mit.edu/mpg/haploview/) (Barrett et al., 2005).
Association analysis
Single-SNP models
The unified mixed linear model in TASSEL 2.1 (Bradbury et al., 2007) was used to identify single-SNP associations in the association population. This model uses the values of estimated membership probability (Q) and pairwise kinship (K) to represent the effects of population structure and relatedness among individuals for marker–trait associations. The Q matrix was identified based on the pattern of population structure (K=3) within the association population (435 unrelated individuals), using 20 neutral simple sequence repeat markers with STRUCTURE v2.3.1 (http://pritchardlab.stanford.edu/structure.html) (Pritchard and Donnelly, 2001). The K matrix was obtained based on the same 20 simple sequence repeat loci using the method proposed by Ritland (1996) in the program SPAGeDi, Ver. 1.2 (Hardy and Vekemans, 2002).
Haplotype-based association models
Haplotype frequencies were estimated from genotype data and haplotype-based association tests with growth and wood properties of P. tomentosa were conducted using the haplotype trend regression software (Zaykin et al., 2002).
Multi-SNP additive and dominance models
Functional genome-wide association studies (fGWAS, version 2.0, http://statgen.psu.edu/software/fgwas-soft.html) was used for identifying and estimating the possible additive and dominant effects associated with all significant SNPs for each trait and was run in R (http://www.r-project.org/). The details of this model have been described by Li et al. (2011).
Multi-SNP epistasis models
Overall architecture of epistatic SNP pairs was obtained by performing FastEpistasis (Schüpbach et al., 2010), an efficient parallel solution extending the PLINK epistasis module, designed to test for SNP–SNP epistasis when analyzing continuous quantitative traits. For much more significant epistatic SNP markers and pairwise SNP interactions, we carried out Multifactor Dimensionality Reduction 3.0.2 (MDR3.0.2) (http://www.multifactordimensionalityreduction.org/) (Hahn et al., 2003).
Results
Expression-based selection and isolation of the Pto-miR257 gene and 12 targets of the Pto-miR257 miRNA
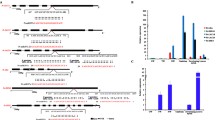
High-throughput RNA sequencing analysis of P. tomentosa miRNAs from three different tissues, along with real-time quantitative of nine diverse tissues, showed that Pto-miR257 was highly expressed in the mature xylem (Figure 1a and Supplementary Data S2), indicating a potentially important role in tree growth and wood formation. Assessment of the secondary structure of Pto-miR257 precursor sequence revealed a typical hairpin structure, confirming Pto-miR257 is a miRNA (Figure 2a). We further identified 97 candidate target genes of Pto-miR257 and selected 12 for further study as they showed high abundance or differential expression in stem (Figures 1a and 2b and Supplementary Data S2). The Pearson correlation coefficients between Pto-miR257 and the 12 targets ranged from −0.808 to −0.510, indicating strong negative correlations of the miRNA and its putative targets. Sixteen gene–gene correlations were also identified for the targets (P⩽0.05, Supplementary Table S3). Subsequently, a correlation network based on Pearson correlation coefficient was constructed (Figure 1b). Moreover, results from degradome sequencing identified the most likely cleavage sites between Pto-miR257 and the target mRNAs (Figure 2c). Then, PCR amplification was performed to obtain the genomic sequence of Pto-miR257 and the 12 target genes: PtoDUF248-1, PtoDUF248-2, PtoAGR1, PtoGH9C1, PtoCKX6, PtoPSKR2-1, PtoPSKR2-2, PtoRBK1-like, PtoFBT8, PtoSepR, PtoDUF620 and PtoAcot13-like.
Expression profiling and a correlation network of Pto-miR257 and its 12 target genes. (a) Relative transcript levels of Pto-miR257 and its targets in various tissues and organs from P. tomentosa. (b) A gene–gene correlation network based on Pto-miR257 targets and highly interrelated (P⩽0.05) target gene pairs was constructed. Solid lines represent positive correlations and dashed lines represent negative correlations; the thickness of the line indicates the strength of correlations, with thicker lines representing more significant correlations.
Secondary structures with or without SNPs in the pre-mature region of Pto-miR257 and possible cleavage sites in the 12 target genes. (a) The 167-nucleotide (nt) pre-mature sequence was used to predict the secondary structure of Pto-miR257. The far left structure represents the hairpin structure of Pto-miR257 without SNPs in the pre-mature region with the mature sequence marked by a black arrow; the others show alterations of secondary structure, along with the free energy of the thermodynamic ensemble (kcal mol−1) changes caused by SNPs in the pre-mature sequence. Red arrows indicate the locus that harbors the allelic variations and SNPs are marked by blue arrows. (b) Target regions and binding sites between Pto-miR257 and its 12 target genes, as determined by psRNATarget; allelic variation within the target sites of PtoPSKR2-1, PtoRBK1-like and PtoSepR also affect miRNA binding with a change of UPE (allowed maximum energy to unpair the target site, kcal mol−1). (c) The most likely cleavage sites between Pto-miR257 and the 12 target genes identified by degradome sequencing.
SNP discovery and nucleotide diversity
We found abundant allelic diversity within Pto-miR257 and its 12 targets. In total, 61 SNPs were detected in Pto-miR257, with an average density of one SNP per 16 bp (πT=0.01680 and θw=0.00480) (Supplementary Table S4). The most conserved region was the mature region, in which only one SNP was identified, where πT=0.00299 and θw=0.00180, respectively. In addition, we identified 8 SNPs in the pre-mature region and 53 SNPs in the flanking region of Pto-miR257, a frequency of 1/21 bp (πT=0.01930 and θw=0.01020) and 1/15 bp (πT=0.00106 and θw=0.01581), respectively. Of these SNPs, we found 58 (95.1%) as common (frequency >10%).
In the 12 potential target genes, we detected 1029 SNPs in total (25 bp−1, πT=0.00618 and θw=0.00626), with 21 to 185 SNPs per gene (Supplementary Tables S4 and S5). All together, 45.6% (469) of these SNPs were considered as common sites (frequency >10%). Analysis of SNP distribution over different regions was conducted and the results are presented in Supplementary Table S5. For each target of Pto-miR257, the SNP frequency was lower in the exons (33 for nonsynonymous sites and 385 for synonymous sites, with a Ka/Ks ratio of 0.09) than in other regions, consistent with the expectation that the coding regions are under stronger selection pressure (Supplementary Tables S4 and S5). The lowest level of nucleotide polymorphism (87 bp−1, 3 variations with 261 bp) was found in the target sites, indicating the target sites show high conservation.
Effects of SNPs on Pto-miR257 biogenesis and target binding
In the present study, we detected 6 common SNPs in the pre-mature region of Pto-miR257 that resulted in 8 different miRNA secondary structures with the free energies of the thermodynamic ensemble of pre-mature sequence ranging from −46.50 to −40.34 kcal mol−1 (Figure 2a). Allelic variation in the miRNA mature region may dramatically change its target spectrum. In this study, we identified one SNP in the mature region of Pto-miR257 (Pto-miR257-SNP32, C>T), we found psRNATarget predicted that the variant miRNA (T allele) no longer targeted 49.5% (48) of the targets of the wild-type Pto-miR257 (C allele), including four candidates: PtoGH9C1, PtoCKX6, PtoSepR and PtoAcot13-like. In addition, psRNATarget also identified 44 new putative targets of this variant miRNA (Supplementary Table S6).
Similarly, SNPs distributed in the targets sites could also affect miRNA binding; as shown in our study, SNPs in miRNA–target binding region of PtoPSKR2-1 (PtoPSKR2-1-SNP7, G>A) and PtoRBK1-like (PtoRBK1-like-SNP51,T>C) both resulted in a change of UPE (allowed maximum energy to unpair the target site) between Pto-miR257 and the respective target mRNA (Figure 2b). Taking PtoPSKR2-1 as an example, UPE was higher for the G allele (19.285 kcal mol−1) than the A allele (18.229 kcal mol−1). The allelic variation of G to C (PtoSepR-SNP30) in the PtoSepR target site also produced a predicted loss of miRNA binding.
SNP genotyping and LD analysis
We selected 527 common SNPs from Pto-miR257 and its 12 targets for genotyping across 435 individuals in the association population and successfully genotyped 496 common SNPs (57 for Pto-miR257 and 439 for the targets) (Supplementary Data S3). The assignment of these loci to different regions within Pto-miR257 and its 12 targets is shown in Supplementary Data S4. Of the genotyped SNPs sites, 51 (89.4%) and 95 (21.6%) were distributed in the flanking region of Pto-miR257 and noncoding regions (5′ and 3′ untranslated regions) of targets, respectively, that harbored the most abundant SNPs compared with other regions of Pto-miR257 and its targets.
We used genotype data for LD analysis and found the overall LD for each gene was low across the sequenced regions with a few loci in strong LD. This analysis also identified several distinct haplotype blocks within Pto-miR257 and its targets (Supplementary Figure S1). For every gene, LD between the SNPs was high within each block (r2 >0.8); nevertheless, LD was mostly low between the haplotype blocks (r2 <0.3). Moreover, we calculated the patterns of LD decay of Pto-miR257 and the 12 targets. The nonlinear regression showed rapid LD decay with distance within each gene (r2 ⩾0.1, within 100 to 900 bp, Supplementary Figure S2), indicating LD of the SNP loci did not extend over the entire gene region.
Single-SNP-based associations
In this study, we performed 4960 association tests between 496 SNPs from Pto-miR257 and its 12 targets and 10 growth and wood traits; these tests identified 62 significant associations representing 44 unique SNPs in Pto-miR257 and its target genes (P<0.01, Q⩽0.10). The percentage of phenotypic variance (R2) explained by individual SNP ranged from 1.2 to 14.2% (Supplementary Table S7).
Of these markers, four (Pto-miR257-SNP3, Pto-miR257-SNP30, Pto-miR257-SNP47 and Pto-miR257-SNP54) from Pto-miR257 were significantly associated with five traits: α-cellulose, hemicellulose, lignin, MFA and V. Markedly, Pto-miR257-SNP30, a SNP in the pre-mature region of Pto-miR257, was significantly associated with α-cellulose content and explained 5.8% of the phenotypic variance (Supplementary Table S7). Measurement of differential transcript abundance across three genotypic classes of the four SNPs indicated that Pto-miR257-SNP30 and Pto-miR257-SNP54 exhibited significant differences in the miRNA transcript levels among different genotypes (Figure 3a).
Transcript analysis of SNP genotypes for significant markers identified by single-SNP-based associations and haplotype/single-SNP-based associations with α-cellulose content and holocellulose content for Pto-miR257 and PtoGH9C1. (a) Transcript abundance of Pto-miR257 and its targets among different genotypic classes. (b) The genotypic effect of the significant haplotype Pto-miR257-SNP30-32 and PtoGH9C1-SNP2-7, along with the genotypic effect for the single marker Pto-miR257-SNP32 and PtoGH9C1-SNP6 in the association population.
For the 439 SNPs from 12 target genes, we found 40 unique SNPs significantly associated with 7 traits, with R2 explained ranging from 1.2 to 14.2%, and 1 to 9 significant loci per gene. For each trait, the number of SNPs ranged from 5 to 13 and most of the significant markers were shared among traits. Of these 40 unique SNPs, there were 7 nonsynonymous, 16 synonymous and 17 noncoding SNPs (Supplementary Table S7). Transcript analysis of SNP genotypes of the 40 significant markers revealed 14 showed differences in the expression level across 3 genotypes and, of these, PtoAGR1-SNP7 exhibited the most significant differences (Figure 3a).
Haplotype-based associations
We also conducted an association test using haplotype trend regression that identified 229 common haplotypes (frequency ⩾10%) from 59 high-LD blocks (r2 ⩾0.7) within Pto-miR257 and its 12 targets. Each block had 2 to 8 common haplotypes, with an average of 4.0 (Table 1); also, each gene had 1 to 10 LD blocks, and 7 to 30 common haplotypes (Table 1). Haplotype-based association detected 75 associations (P⩽0.01, Q⩽0.10, frequency ⩾10%) representing 50 haplotypes from 28 blocks within 10 genes (except PtoAGR1 and PtoAcot13-like) that associated with 9 traits (Table 1 and Supplementary Table S8). Each trait had 4 to 18 significant haplotypes and most of the haplotypes simultaneously associated with several traits, indicating pleiotropic effects of important loci. For instance, one haplotype (T-C-G-C-G) in block 1 from SNP24 to SNP28 in PtoDUF620 showed a close association with α-cellulose, holocellulose and lignin contents. In addition, many haplotype-based associations were strongly supported by significant single-marker associations (Supplementary Tables S7 and S8).
Multi-SNP associations under additive and dominant effects model
We employed Bayesian hierarchical models that emphasized multi-SNP additive and dominant effects for each quantitative trait to identify new genetic associations, including 97 significant marker–trait pairs for 71 unique SNPs from Pto-miR257 and its 12 targets associated with all 10 growth and wood property traits, with R2 of 0.5 to 8.5% (average R2=3.0%) (Supplementary Table S9). Overall, we observed 89.1% of the associations involved a combination of additive and dominant effects. The total numbers of identified SNP–trait associations varied across trait categories: the 97 significant associations included 24 for wood chemical compositions, 38 for wood physical properties and 35 for growth traits (Supplementary Table S9). Three SNPs (PtoPSKR2-2-SNP12, PtoSepR-SNP27 and PtoAcot13-like-SNP24) were uniquely examined in this case, compared with single-SNP-based associations. Moreover, this algorithm also uncovered numerous markers that were not obtained using the single-marker model, including PtoPSKR2-1-SNP7 and PtoSepR-SNP30 in the binding sites of Pto-miR257 targets (Supplementary Tables S7 and S9).
Additive effect
We detected 92 significant associations under the additive effect model, involving 69 unique SNPs from Pto-miR257 and its 12 targets and 10 traits (Table 2 and Supplementary Table S9). Of these, 19 loci showed significant associations with at least 2 traits, and 9 of the loci were from Pto-miR257, indicating the important role of Pto-miR257 in wood formation. Correspondingly, each trait was associated with variation in at least two candidate genes and 2 to 14 significant, unique SNPs. In the present study, we found Pto-miR257 was significantly associated with 7 traits, with an average additive effect of 1.480. All the target genes, except PtoDUF248-2 and PtoRBK1-like, were also associated across all three growth traits.
Dominant effect
Under the dominance model, we detected 92 significant associations involving 68 unique SNPs from Pto-miR257 and its 12 targets and 10 traits, with dominance effects of −4.772 to 4.846 (Table 3 and Supplementary Table S9). Of these, 46 had positive dominance values and 46 had negative values. The number of significant associations with positive versus negative values across the three trait categories was 20/17 for wood physical properties, 12/8 for wood chemical compositions and 14/21 for growth traits. In addition, 10 SNPs from 5 genes simultaneously exhibited opposite dominant values for different traits. Most candidates (10) showed dominant effects within or across trait categories, and we identified different SNPs with different effects for the same gene.
Multi-SNP associations under the epistatic model
By performing the FastEpistasis, we identified 2722 SNP–SNP epistasis involving 369 SNPs within Pto-miR257 and its 12 target genes that associated with 10 traits (P<1E−03, Supplementary Data S5 and S6). In addition, 12.6% (48) of the significant SNPs were simultaneously examined under the additive and dominant effect models (Supplementary Table S9 and Supplementary Data S5), indicating the complicated genetic mechanism by which allelic variation contributes to phenotypic variation. Furthermore, internal networks were constructed according to these significant allelic epistatic effects, reflecting the interactions of miRNA–mRNA and mRNA–mRNA pairs (Figure 4a). Here, we found epistatic effects widely occurred among different gene pairs, revealing that the genetic architecture of complex quantitative traits is highly dynamic and sensitive to the exact allelic composition (Figure 4a). Moreover, we observed most genes, including Pto-miR257, have a combination of three main effects within a complex genetic network (Table 4 and Figure 4a).
Epistatic interactions among Pto-miR257 and its target genes for α-cellulose content and DBH and transcript analysis of significant SNP genotypes from Pto-miR257 and PtoDUF248-1. (a) Networks of gene–gene epistatic interactions. Thicker lines represent interactions between Pto-miR257 and its targets and thinner lines represent target–target epistatic effects; dark shading indicates genes harboring significant SNPs that hold a combination of three hereditary effects (additive, dominant and epistasis). (b) Entropy-based interaction graph of target-target epistatic interactions for PtoDUF248-1, PtoGH9C1, PtoPSKR2-1, PtoSepR and miRNA–target interactions for Pto-miR257, PtoDUF248-1, PtoRBK1-like and PtoFBT8. Values in the boxes represent the individual information gain of one single SNP; the positive/negative values along the line indicate positive/negative effects, respectively. Red and blue lines indicate candidates that were further explored for different genotype combinations affecting epistatic interactions and transcript abundance in (c) and (d) below, respectively. (c) Epistatic effects of different genotype combinations for candidate genes analyzed in (b) above. Dark/light shaded groups represent high/low interaction values and the vertical lines/boxes indicate the higher/lower interaction effects of different genotype combinations than the mean values of the three separate genotypes. DBH, diameter at breast height. (d) Transcript levels of PtoDUF248-1 with nine genotypic classes from Pto-miR257-SNP56 and PtoDUF248-1-SNP33. CC, CT and TT and GG, GC and CC were observed in the locus of Pto-miR257-SNP56 and PtoDUF248-1-SNP33, respectively.
We next examined the most remarkable allelic interactions for each trait using MDR 3.0.2 and this validated some of the SNP–SNP pairs identified previously. This algorithm detected 55 associations across 10 traits, including 49 unique SNPs from Pto-miR257 and its 11 target genes (except PtoAcot13-like), with main effects of 0.01 to 4.96% (Supplementary Table S10). After analyzing the pairwise effects, we identified 110 significant SNP-SNP pairs with epistatic interactions of −8.47 to 2.44% (Supplementary Table S11). Of the total SNP–SNP interactions, more than half (56) represented miRNA–mRNA interactions, with epistatic effects of −8.47 to 2.09%. Moreover, negative interactions existed between Pto-miR257 and 11 of its targets, which reflected redundancy between loci, indicating these loci provide, in part, the same information for these traits (Hahn et al., 2003). We also identified negative epistatic effects between two targets that encode the same protein, PtoDUF248-1 and PtoDUF248-2. In contrast, we only found 18.2% (20) of the SNP pairs showed positive interactions, representing jointly higher effects than single SNPs. Considering the additive and dominant effects together, we noticed two SNP markers, Pto-miR257-SNP56 and PtoDUF248-1-SNP21, show a combination of these three effects (Supplementary Tables S9 and S10).
To visualize the two-way interaction for traits, we created an entropy-based interaction graph for α-cellulose content and DBH that indicated interactions between seven SNPs in Pto-miR257, PtoDUF248-1, PtoGH9C1, PtoPSKR2-1, PtoRBK1-like, PtoFBT8 and PtoSepR (Figure 4b). Our analysis also demonstrated allele epistatic interactions existed not only between Pto-miR257 and its target genes (miRNA-mRNA), but also among its targets (mRNA–mRNA). Figure 4c exhibits the genotypic combinations associated with high and low epistatic values for SNP markers involved in Figure 4b. The patterns of high- and low-value groups that clearly differ across each of the different multilocus dimensions were considered as evidence of epistasis, or gene–gene interactions.
Discussion
Allelic SNPs affecting miRNA biogenesis and target accessibility
SNPs in the pre-mature region of miRNA genes could affect miRNA abundance by affecting the stability of the hairpin structure, and this can contribute to miRNA-mediated translational suppression and phenotypic variation (Yang et al., 2014). Here we detected six SNPs in the pre-mature region of Pto-miR257, resulting in eight different secondary structures with various free energies of the thermodynamic ensemble (Figure 2a). Notable, analysis of transcript levels found Pto-miR257-SNP30, one of the six SNPs, exhibited significant differences among the three genotypes (Figure 3a). In addition, association studies found it showed a strong association with α-cellulose content, both by single-SNP and haplotype-based association analysis (Figure 3b). Our study further validated the idea that SNPs in miRNA genes may change the production of the mature miRNAs and contribute to genetic and phenotypic variation. The miRNAs recognize their target mRNAs mainly by base pairing between nucleotides within the mature region and the complementary nucleotides on the target mRNAs, and hence allelic variants in mature domains could directly influence miRNA target binding and selection. Our study found the specific variation in Pto-miR257-SNP32 would cause loss and gain of roughly half of its targets (Supplementary Table S6), supporting the idea that SNPs in mature region of the miRNA will alter the target profile by causing the loss of original targets or gain of new targets (Liu et al., 2013). Here, we did not consider the variant Pto-miR257 (T) and wild-type miRNA (C) as isoforms within the same miRNA family as this variation (Pto-miR257-SNP32, C>T) was just a single-base mutation that happened at the same locus of precursor region and did not alter the secondary structure of Pto-miR257 (Supplementary Figure S3). Furthermore, we searched against the other novel miRNAs identified by high-throughput RNA sequencing, along with the conserved miRNAs deposited at miRBase 21.0 (http://www.mirbase.org/index.shtml) and did not find the variant miRNA.
As miRNA-mediated regulation mainly relies on the interaction of the miRNA and its target site, SNPs within the binding sites of target genes probably disrupt sequence complementarity and change the binding affinity, possibly producing phenotypic variation. In our study, Pto-miR257 had a higher predicted binding affinity for the G allele of PtoPSKR2-1 than the A allele (Figure 2b). The miRNA target loss also happened owing to the G to C allele in the PtoSepR target site. Consistent with alterations in binding affinity affecting regulation, SNPs in binding regions of Pto-miR257 target genes were also associated with regulation and phenotypic variance. We found by performing haplotype-based association analysis that PtoPSKR2-1-SNP7 was part of a haplotype closely linked to FW, D and V (Supplementary Table S8). Moreover, multi-SNP association analysis revealed this haplotype also showed a combination of three effects. Meanwhile, another marker, PtoSepR-SNP30, also showed three genetic effects. Our study thus demonstrates target accessibility is a critical factor in miRNA function and may govern miRNA-mediated gene regulation.
SNPs within Pto-miR257 and its target genes associated with growth and wood properties
Through performing single-SNP association analysis, we identified 62 significant associations within Pto-miR257 and its targets (Supplementary Table S7). We are gratified to find that Pto-miR257 was simultaneously associated with three categories of traits (average R2=6.2%); for its targets, we observed most of the significant markers were shared among traits. Our finding hence supported the idea that Pto-miR257 and the 12 targets likely participate in wood biosynthesis and tree growth.
Under the haplotype-based association model, we detected 50 significant haplotypes involving 75 associations (Supplementary Table S8). Many of the identified haplotype-based associations were strongly supported by single-marker associations that always had higher R2 than other haplotypes, probably because the significant markers in these haplotypes were closely linked to the same trait (Supplementary Tables S7 and S8). For instance, PtoGH9C1-SNP6 (A>G), which was significantly associated with holocellulose content by single-SNP association, also had a significant haplotype (A-A-G-T-C-C) that showed a strong association with the same trait and explained 19.4% of the phenotypic variance (Figure 3b). In rice, OsGH9C1 is a member of the OsGH9 family that takes part in the modification of cellulose crystallinity (Xie et al., 2013). In poplar, PtoGH9C1 may participate in the biosynthesis of cellulose, as it is a paralog of OsGH9C1. Our results agreed with this deduction and the high abundance of PtoGH9C1 in developing xylem provides more supporting evidence (Figure 1a).
By multi-SNP association analysis, we found that SNPs in 10 targets (except PtoAGR1 and PtoPSKR2-2) shared at least one common trait with Pto-miR257, reflecting the possible genetic interaction between Pto-miR257 and its target genes and indicating that they may function in the same pathway. For the targets, three missense mutations (PtoPSKR2-2-SNP12, PtoSepR-SNP27 and PtoAcot13-like-SNP24) that cause amino acid changes from Ser to Pro, Asp to His and Thr to Ser, respectively, were associated with the same trait (holocellulose, hemicellulose and α-cellulose contents, respectively), compared with significant markers identified by single-SNP-based associations, emphasizing the potential function of the three targets in cellulose synthesis (Supplementary Tables S7 and S9). The high abundance of their transcripts in cambium supports the potential crucial role they play in wood formation (Figure 1a). Our observations thus demonstrated that nonsynonymous substitutions within genes, along with variations of encoded amino acids, could alter the carrier’s phenotype, probably because of their marked effects on structure and functional properties of proteins. Moreover, our study also improved our understanding of PtoPSKR2-2, PtoSepR and PtoAcot13-like (Wei et al., 2009; Mosher and Kemmerling, 2013; Zhang et al., 2014).
Furthermore, this algorithm (Bayesian hierarchical models) examined two target genes, PtoDUF248-1 and PtoDUF248-2, predicted to encode proteins with putative methyltransferase (MTase) function, both significantly associated with hemicellulose and lignin content. Previous studies have verified the function of these proteins in methylesterification of the pectin homogalacturonan (Miao et al., 2011), showing that these two targets play crucial roles in cell wall formation. We were curious whether they also participate in biosynthesis of other factors that affect cell wall development, such as lignin and cellulose. Taking the significant single-SNP associations, we observed these two targets were closely linked to α-cellulose content. In addition, real-time quantitative PCR showed both of them had high expression in cambium (Figure 1a). All of these findings support the idea that PtoDUF248-1 and PtoDUF248-2 play significant roles in cell wall biosynthesis, not only via their function in pectin composition, but also via their potential effects on lignocellulose synthesis.
Plant hormones contribute to diverse processes in tree growth (Shen, 1996). Two of the targets function in hormone-mediated processes: PtoAGR1 affects auxin transport (Shin et al., 2005) and PtoCKX6 affects the degradation of cytokinins (Schmülling et al., 2003). In this study, we detected significant markers in PtoAGR1 that were closely linked to holocellulose content, and prominent SNPs belonging to PtoCKX6 that associated with MFA, H and V (Supplementary Table S9). Our findings demonstrate their potential role in cellulose formation and tree growth, providing us with a new understanding of their function. When taking into account their transcript patterns in different tissues of P. tomentosa, we observed both of them showed high expression in xylem and shoot apex (Figure 1a), further proving their important role in tree growth. However, the effects of correlative factors in wood development remain unknown and will be interesting to explore in the future. Thus, our study successfully identified several candidates that may have significant functions in wood biosynthesis and tree growth, including Pto-miR257 and its 12 target genes, that may prove useful for poplar breeding.
Epistasis effects reflect the miRNA–mRNA interactions
In epistasis, an allele of one gene masks the phenotype of an allele of another gene, and this can provide us with information complementary to that gained through single-locus analysis. Reports on human diseases revealed epistasis played significant roles in disease susceptibility, thus suggesting complex genetic interactions may have stronger effects than any single gene (Chou et al., 2011). In addition, there is evidence that epistatic interactions between quantitative trait loci could contribute to mean genotypic values in Drosophila and Arabidopsis (Mackay, 2014; Lovell et al., 2015). Thus, epistasis analysis has considerable potential to reveal interactions between genes, including regulatory pathways between a miRNA and its targets, and their effects on complex traits of trees.
Here, we used FastEpistasis and MDR 3.0.2 to examine SNP–SNP epistasis for Pto-miR257 and its target genes associated with the phenotypic traits. We observed epistasis between miRNA–mRNA and mRNA–mRNA pairs (Supplementary Table S11 and Supplementary Data S6). Importantly, most of the SNPs belong to miRNA–mRNA pairs, supporting the interaction between Pto-miR257 and its target genes. Interconnected networks reflecting gene–gene epistatic interactions, along with entropy-based interaction graphs mirroring two-way interactions of miRNA–mRNA and mRNA–mRNA pairs, were constructed in this study (Figure 4). In addition, deriving genetic interaction networks from epistatic interactions between loci will improve our understanding of biological systems that give rise to variation in quantitative traits. To dissect the epistatic effects between Pto-miR257 and its targets, we also calculated epistatic values of different genotype combinations (Figure 4c). After transcript-level analysis, we found PtoDUF248-1 exhibited no significant differences with different genotype combinations of Pto-miR257-SNP56 and PtoDUF248-1-SNP33 when the genotype of Pto-miR257 was CC or CT. This indicates that C allele prevents the variation in PtoDUF248-1-SNP33 from manifesting its effects, and this may reflect functional interaction of miRNA–targets (Figure 4d).
The MDR 3.0.2 model can work out epistasis effects with high/low values owing to different genotype combinations that may provide a new resource for exploring the interactions underlying miRNA–target effects. With this model, we found negative epistatic interactions occurred between Pto-miR257 and its 11 targets, consistent with their opposite expression patterns (Figure 1a) and reflecting negative regulatory relationships between them. Genes interacting via negative epistatic interactions often carry out related roles where the absence of one can be compensated for by the other; our finding hence suggested Pto-miR257 and its target genes may have similar roles and function in the same pathway. We found the SNP pairs, Pto-miR257-SNP10 and PtoGH9C1-SNP16, that were significantly associated with α-cellulose content. This observation further demonstrates PtoGH9C1 has potential effects on lignocellulose synthesis (Xie et al., 2013).
Here, we detected significant epistatic interactions between Pto-miR257 and its targets and identified negative epistatic values among them, reflecting the functional miRNA–mRNA interaction. Epistatic values in target–target interactions, indicating interactions between target genes, will also influence the biological process of tree growth. However, the detailed regulatory mechanisms underlying them are still unknown and thus require further research. Furthermore, the present work found that Pto-miR257 and most of its targets were significantly associated with one trait at least two times, demonstrating their effect on related phenotypes. In addition, at least two such genes contributed to one trait, indicating tree growth and wood biosynthesis require the coordinate regulation of diverse genes that function in shared or different metabolic pathways (Table 4). Our work verified the importance of epistasis as a main factor that affects phenotypic traits and provides a strategy for exploring genetic networks related to these traits.
Conclusions
In this study, we used SNP-based association analysis to explore the function of Pto-miR257 and 12 of its target genes, and the interactions of the miRNA with its targets. We aimed to investigate the role of Pto-miR257, target genes and miRNA-mediated negative regulation in wood biosynthesis and tree growth. Our study thus demonstrates that these genes act as significant factors affecting wood formation. Here we combined analysis of transcript levels, degradome sequencing, single-SNP and haplotype-based association analysis and multi-SNP association analysis, taking into account additive, dominant and epistatic effects to examine the function and interactions of miRNAs with their target genes. The results showed negative regulation and dynamic relations mirrored by epistatic effects between Pto-miR257 and its target genes. Moreover, epistasis can be used to define functional relationships between genes, and to identify genetic regulatory networks of miRNAs–targets. This work thus contributes to improving our understanding of the mechanisms of gene regulation and phenotypic variance that occurs based on miRNA–target interactions that could be helpful for miRNA studies in biology.
References
Barrett JC, Fry B, Maller J, Daly MJ . (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265.
Beaulieu J, Doerksen T, Boyle B, Clément S, Deslauriers M, Beauseigle S et al. (2011). Association genetics of wood physical traits in the conifer white spruce and relationships with gene expression. Genetics 188: 197–214.
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES . (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635.
Chou HH, Chiu HC, Delaney NF, Segre' D, Marx CJ . (2011). Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332: 1190–1192.
Dai XB, Zhao PX . (2011). psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39: 155–159.
Dusl M, Senderek J, Müller JS, Vogel JG, Pertl A, Stucka R et al. (2015). A 3'-UTR mutation creates a microRNA target site in the GFPT1 gene of patients with congenital myasthenic syndrome. Hum Mol Genet 24: 3418–3426.
Du QZ, Xu BH, Gong CR, Yang XH, Pan W, Tian JX et al. (2014). Variation in growth, leaf, and wood property traits of Chinese white poplar (Populus tomentosa, a major industrial tree species in Northern China. Can J Forest Res 44: 326–339.
Hahn LW, Ritchie MD, Moore JH . (2003). Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics 19: 376–382.
Hardy OJ, Vekemans X . (2002). Spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2: 618–620.
Hill WG, Robertson A . (1968). Linkage disequilibrium in finite populations. Theor Appl Genet 38: 226–231.
Kwon E, Wang W, Tsai LH . (2013). Validation of schizophrenia-associated genes CSMD1 C10orf26 CACNA1C and TCF4 as miR-137 targets. Mol Psychiatry 18: 11–12.
Lei B, Gao S, Luo LF, Xia XY, Jiang SW, Deng CY et al. (2011). A SNP in the miR-27a gene is associated with litter size in pigs. Mol Biol Rep 38: 3725–3729.
Li JH, Kiranmoy D, Fu GF, Li RZ, Wu RL . (2011). The Bayesian lasso for genome-wide association studies. Bioinformatics 27: 516–523.
Liu QP, Wang H, Zhu LY, Hu HC, Sun YQ . (2013). Genome-wide identification and analysis of miRNA-related single nucleotide polymorphisms (SNPs) in rice. Rice 6: 10.
Lovell JT, Mullen JL, Lowry DB, Awole K, Richards JH, Sen S et al. (2015). Exploiting differential gene expression and epistasis to discover candidate genes for drought-associated QTLs in Arabidopsis thaliana. Plant Cell 27: 969–983.
Lu SF, Li QZ, Wei HR, Chang MJ, Tunlaya-Anukit S, Kim H et al. (2013). Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proc Natl Acad Sci USA 110: 10848–10853.
Mackay TF . (2014). Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat Rev Genet 15: 22–33.
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL et al. (2008). Criteria for annotation of plant microRNAs. Plant Cell 20: 3186–3190.
Miao Y, Li HY, Shen J, Wang J, Jiang L . (2011). QUASIMODO3 QUA3 is a putative homogalacturonan methyltransferase regulating cell wall biosynthesis in Arabidopsis suspension-cultured cells. J Exp Bot 62: 5063–5078.
Mosher S, Kemmerling B . (2013). PSKR1 and PSY1R-mediated regulation of plant defense responses. Plant Sign Behav 8: e24119.
Pritchard JK, Donnelly P . (2001). Case-control studies of association in structured or admixed populations. Theor Popul Biol 60: 227–237.
Ritland K . (1996). Estimators for pairwise relatedness and individual inbreeding coefficients. Genet Res Camb 67: 175–185.
Robischon M, Du J, Miura E, Groover A . (2011). The Populus class III HD ZIP, popREVOLUTA, influences cambium initiation and patterning of woody stems. Plant Physiol 155: 1214–1225.
Rogers K, Chen XM . (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399.
Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R . (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19: 2496–2497.
Schmülling T, Werner T, Riefler M, Krupková E, Manns IB . (2003). Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J Plant Res 116: 241–252.
Schüpbach T, Xenarios I., Bergmann S, Kapur K . (2010). FastEpistasis: a high performance computing solution for quantitative trait epistasis. Bioinformatics 26: 1468–1469.
Shen HJ . (1996). Plant hormone and wood formation. Sci Silwae Sin 32: 165–170.
Shin H, Shin H, Guo ZB, Blancaflor EB, Masson PH, Rujin Chen RJ . (2005). Complex regulation of Arabidopsis AGR1/PIN2-mediated root gravitropic response and basipetal auxin transport by cantharidin-sensitive protein phosphatases. Plant J 42: 188–200.
Song YP, Ma KF, Ci D, Zhang ZY, Zhang DQ . (2013). Sexual dimorphism floral microRNA profiling and target gene expression in andromonoecious poplar Populus tomentosa. PLoS One 8: e62681.
Sun K, Lai EC . (2013). Adult-specific functions of animal microRNAs. Nat Rev Genet 14: 535–548.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S . (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739.
Thumma BR, Nolan MF, Evans R, Moran GF . (2005). Polymorphisms in cinnamoyl CoA reductase CCR are associated with variation in microfibril angle in Eucalyptus spp. Genetics 171: 1257–1265.
Wei J, Kang HW, Cohen DE . (2009). Thioesterase superfamily member 2 Them2/acyl-CoA thioesterase 13 Acot13: a homotetrameric hotdog fold thioesterase with selectivity for long-chain fatty acyl-CoAs. Biochem J 421: 311–322.
Xie GS, Yang B, Xu ZD, Li FC, Guo K, Zhang ML et al. (2013). Global identification of multiple OsGH9 family members and their involvement in cellulose crystallinity modification in rice. PLoS One 8: e50171.
Yang XH, Du QZ, Chen JH, Wang BW, Zhang DQ . (2015). Association mapping in Populus reveals the interaction between Pto-miR530a and its target Pto-KNAT1. Planta 242: 77–95.
Yang Q, Jie Z, Ye S, Li Z, Han Z, Wu J et al. (2014). Genetic variations in miR-27a gene decrease mature miR-27a level and reduce gastric cancer susceptibility. Oncogene 33: 193–202.
Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG . (2002). Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered 53: 79–91.
Zhang DQ, Du QZ, Xu BH, Zhang ZY, Li BL . (2010). The actin multigene family in Populus: organization, expression and phylogenetic analysis. Mol Genet Genomics 284: 105–119.
Zhang YP, Nandakumar R, Bartelt-Hunt SL, Snow DD, Hodges L, Li X . (2014). Quantitative proteomic analysis of the Salmonella-lettuce interaction. Microb Biotechnol 7: 630–637.
Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities (No. TD2012-01), National ‘863’ Plan Project (No. 2013AA102702) and the Project of the National Natural Science Foundation of China (No. 31170622).
Author contributions
DZ designed the research; BC, QD, JC, XY, JT and DZ performed research; BC analyzed and interpreted the data; and BC wrote the paper. All authors commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on Heredity website
Supplementary information
Rights and permissions
About this article
Cite this article
Chen, B., Du, Q., Chen, J. et al. Dissection of allelic interactions among Pto-miR257 and its targets and their effects on growth and wood properties in Populus. Heredity 117, 73–83 (2016). https://doi.org/10.1038/hdy.2016.26
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2016.26
- Springer Nature Switzerland AG