Abstract
The chemokine CXCL10 and its receptor CXCR3 play a role in breast cancer metastasis to bone and osteoclast activation. However, the mechanism of CXCL10/CXCR3-induced intracellular signaling has not been fully investigated. To evaluate CXCL10-induced cellular events in the mouse breast cancer cell line 4T1, we developed a new synthetic CXCR3 antagonist JN-2. In this study, we observed that secretion of CXCL10 in the supernatant of 4T1 cells was gradually increased during cell growth. JN-2 inhibited basal and CXCL10-induced CXCL10 expression and cell motility in 4T1 cells. Treatment of 4T1 cells with CXCL10 increased the expression of P65, a subunit of the NF-κB pathway, via activation of the NF-κB transcriptional activity. Ectopic overexpression of P65 increased CXCL10 secretion and blunted JN-2-induced suppression of CXCL10 secretion, whereas overexpression of IκBα suppressed CXCL10 secretion. These results indicate that the CXCL10/CXCR3 axis creates a positive feedback loop through the canonical NF-κB signaling pathway in 4T1 cells. In addition, treatment of osteoblasts with conditioned medium from JN-2-treated 4T1 cells inhibited the expression of RANKL, a crucial cytokine for osteoclast differentiation, which resulted in an inhibitory effect on osteoclast differentiation in the co-culture system of bone marrow-derived macrophages and osteoblasts. Direct intrafemoral injection of 4T1 cells induced severe bone destruction; however, this effect was suppressed by the CXCR3 antagonist via downregulation of P65 expression in an animal model. Collectively, these results suggest that the CXCL10/CXCR3-mediated NF-κB signaling pathway plays a role in the control of autonomous regulation of CXCL10 and malignant tumor properties in breast cancer 4T1 cells.
Similar content being viewed by others
Introduction
The tumor microenvironment contributes to the malignant features of cancer cells by sustaining tumor growth and metastasis. Bone is well characterized as a preferential metastatic site of breast cancer, and bone metastasis consequentially induces osteolytic lesions through interactions between cancer cells and the bone marrow microenvironment.1, 2 The development of osteolytic bone metastasis is associated with colonization of cancer cells to bone and the production of osteolytic factors by cancer cells, which induces osteoclast-mediated bone resorption and destruction.3, 4 Growth factors from the degraded bone matrix subsequently stimulate tumor growth, resulting in a vicious cycle in bone metastasis. Bone metastasis is an advanced cancer that induces fragile bone, pain and spinal cord compression.5
The multifunctional roles of the chemokine network in tumors are well established.6, 7, 8 Indeed, chemokines were initially characterized not only in leukocyte adhesion and migration under a variety of physiological and pathological conditions but also in hematopoiesis, lymphocyte development, and wound healing.9, 10 However, increasing evidence suggests that chemokines and their receptors play a role in tumor initiation, progression, and metastasis because invasion and metastasis of cancer cells share many similarities with the process of leukocyte infiltration.9, 11, 12 Interactions of chemokines and their receptors, such as CXCL12 with CXCR4, play a critical role in determining the metastatic site of breast cancer for several organs.13, 14 Similarly, the interaction of CXCL10 with CXCR3 also plays an important role in metastasis in various cancer cells, including colorectal carcinoma cells, breast cancer, colon cancer, melanoma and glioma.15 Expression profiles of chemokine receptors indicate that both CXCR3 and CXCR4 are significantly increased in colorectal liver metastases compared with the corresponding primary colorectal cancer.16 Other pairs, such as CCL21/CCR7, are also established in cancer metastasis.17 Therefore, accumulating evidence suggests that the interaction of chemokines and their receptors promote malignant tumor properties.
CXCL10, also known as interferon-gamma-induced protein 10 (IP-10), is well characterized as a chemoattractant for immune cells, such as T-lymphocytes and monocytes, upon activation of its receptor, CXCR3.18 In inflammatory conditions, CXCL10 is secreted from a variety of cells, including monocytes, endothelial cells, fibroblasts and keratinocytes, in response to interferon-gamma.19 In addition, we observed that melanoma cells secrete higher levels of CXCL10 than macrophages.20 Although interferon-gamma is a major inducer of CXCL10 expression, the mechanism by which high expression of CXCL10 is maintained in cancer cells has not been fully elucidated. A previous report observed that CXCL10 induction is driven by tumor necrosis factor α (TNFα)-induced NF-κB transcriptional activation in endothelial cells.21 In microglia cells, CXCL10 expression occurs through the p38/MAPK, JNK/MAPK and NF-κB cascade.22 Indeed, NF-κB binds to the κB2-binding site of the CXCL10 promoter that contains a homodimer of P65.23
In the present study, we aimed to elucidate the action of CXCL10 on CXCR3-mediated NF-κB signaling in breast cancer 4T1 cells. In addition, we evaluated our newly developed CXCR3 antagonist, JN-2, in CXC10/CXCR3-mediated intracellular signaling and assessed its potential in 4T1 cells. Our findings will provide insight into the fundamental role of chemokines in cancer, which will subsequently shed light on which chemokine-mediated cellular events are essential for malignant tumor properties.
Materials and methods
Biological compounds
The CXCR3 antagonist N-(4-(5-chlorobenzo[d]oxazol-2-ylamino)phenyl)-4-aminobutanamide (JN-2) was synthesized and purified as described in the Supplementary Materials. AMG 487 was obtained from Tocris Bioscience (Ellisville, MO, USA).
Reagents and antibodies
Recombinant mouse CXCL10 was obtained from PeproTech EC (London, UK). To detect specific protein expression by western blot, antibodies against P65 (cat. #8242), IκBα (cat. #4812) and α-tubulin (cat. #2144) were purchased from Cell Signaling Technology (Danvers, MA, USA). Antibodies against Lamin B (cat. #sc-6217) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA). The antibody against β-actin (cat. #A5441) was obtained from Sigma-Aldrich (St Louis, MO, USA). Secondary antibodies conjugated with horseradish peroxidase were purchased from Sigma-Aldrich.
Animal experiments
All animal procedures were performed in accordance with the guidelines of the Animal Care Committee of the Institute of Laboratory Animal Resources of Seoul National University (Seoul, Korea). Six-week-old female BALB/c mice (Orient Bio Inc., Seoul, Korea) were intraperitoneally injected with DMSO or JN-2 (10 mg kg−1 of body weight) every other day starting 7 days before 4T1 injection (n=7 per group). Then, 4T1 cells were collected from subconfluent cultures, washed with PBS twice and resuspended in PBS. An amount of 1 × 104 cells in 5 μl PBS was injected into the femoral marrow space through the femoral condyle by using a 30-gauge Hamilton syringe. All mice were killed on day 21 after 4T1 injection. The femurs were flash-frozen in liquid nitrogen and stored at −80 °C for RNA extraction or fixed in 4% paraformaldehyde for micro-computed tomography (μCT) and histological studies.
The bone architecture in the femur was analyzed by high-resolution μCT (SMX-90CT system; Shimadzu, Kyoto, Japan). Scanned images from μCT were reconstructed using the VG Studio MAX 1.2.1 program (Volume Graphics, Heidelberg, Germany). The three-dimensional images were used to measure bone volume, cortical bone volume and trabecular number with the TRI/3D-VIE (RATOC System Engineering, Kyoto, Japan) program.
Histology
Histomorphometric analysis was performed using femurs decalcified with 12% EDTA for 3 weeks and embedded in paraffin. After 5-μm histological sagittal sections were made, femurs were stained with tartrate-resistant acid phosphatase (TRAP) solution (Sigma-Aldrich) or hematoxylin and eosin (H&E) solution (Sigma-Aldrich). Osteoclast number was measured by using the OsteoMeasure XP program (version 1.01; OsteoMetrics, Decatur, GA, USA).
For immunohistological analysis, paraffin-embedded samples were sectioned at 5-μm thickness and mounted on silane-coated slides (Muto-Glass; Tokyo, Japan). Deparaffinization was performed in xylene, a xylene/ethanol mixture and serial ethanol dilutions. For antigen retrieval, sections were incubated with 10 mM citrate (pH 6.0) containing 0.05% Tween-20 for 20 min at 80 °C followed by a PBS wash containing 0.05% Tween-20. Sections were then pre-incubated with blocking buffer containing 5% goat serum, 0.05% sodium azide and 0.1% Triton X-100 in PBS for 45 min at room temperature and incubated overnight at 4 °C with anti-P65 antibody (Cell Signaling Technology) in PBS containing 1% goat serum, 0.05% sodium azide and 0.02% Tween-20. Immunostaining was performed according to the manufacturer’s instructions using the ChemMate Dako Envision Detection kit, Peroxidase/DAB, Rabbit/Mouse (DakoCytomation, Glostrup, Denmark).
Cells and culture system
The 4T1 mouse breast cancer cell line was obtained from the American Type Culture Collection (Manassas, VA, USA) and cultured in RPMI complete media containing 10% heat-inactivated fetal bovine serum, 50 units ml−1 of penicillin and 50 μg ml−1 of streptomycin. Bone marrow cells were obtained by flushing bone marrow from the femur of 5-week-old BALB/c mice, and non-adherent bone marrow cells were cultured with M-CSF (60 ng ml−1) for 3 days to generate bone marrow-derived macrophages (BMMs) as previously described.24 To differentiate osteoclasts, BMMs (4 × 104 cells per well) were cultured in 48-well tissue culture plates with α-minimum essential medium (α-MEM) including M-CSF (60 ng ml−1) and RANKL (100 ng ml−1) for 4 days in 48-well culture dishes. The complete medium was changed on the third day. Osteoclasts were washed with PBS, fixed with 3.7% formalin for 10 min, permeabilized with 0.1% Triton X-100, and then stained for TRAP (Sigma-Aldrich). The co-culture system was performed by culturing BMMs (1 × 105 cells per well) and primary osteoblasts (2 × 104 cells per well) in 48-well (1 ml per well) tissue culture plates for 9 days in α-MEM. At the end of the culture period, cells were stained by TRAP as described above.
Motility assay
A scratch wound healing assay was performed on monolayers of confluent 4T1 cells scraped with a p200 pipet tip, and then the medium was replaced with RPMI complete media containing the indicated dose of DMSO or JN-2 and PBS or mCXCL10. Phase-contrast images were acquired after scratching and after 24 h of incubation at 37 °C. Transwell assays were performed using the upper chamber (8 μm pore membranes; Corning, NY, USA). Briefly, 4T1 cells (5 × 104) were seeded on the transwell upper chamber with RPMI complete media, and the lower chambers were treated with DMSO or JN-2 and PBS or mCXCL10 with RPMI complete media. After 24 h of incubation, unmigrated cells were removed from the top of the upper membranes, and migrated cells were fixed with 3.7% formalin for 10 min and stained with H&E solution for 1 min.
Preparation of conditioned medium and ELISA
Briefly, 4T1 cells (1 × 105 cells per well) were cultured in RPMI complete media on six-well tissue culture plates for 12 h and were then treated with DMSO or JN-2 as indicated. After 24 h of incubation, the supernatant of conditioned medium (CM) was harvested, centrifuged at 2000 r.p.m. for 5 min, and stored at −80 °C. The secreted CXCL10 in the cell culture medium was measured using the corresponding ELISA kit (R&D Systems, Minneapolis, MN, USA). Serum pyridinoline (PYD) was determined with the Metra Serum PYD EIA kit (Quidel Corporation, San Diego, CA, USA).
Cell viability assay
Briefly, 4T1 cells (1 × 104 cells per well) were cultured in RPMI complete media on 96-well culture plates for 12 h, and the media were exchanged with DMSO or the indicated doses of JN-2. After 24 h of incubation, 10 μl of EZ-Cytox solution (Daeillab Service, Seoul, Korea) was added to each well of the plate and incubated at 37 °C for 2 h. Absorbance was measured by spectrophotometry (Mutiskan FC, Thermo Fisher Scientific, Waltham, MA, USA) at 450 nm.
Western blot
For western blot sample preparation, the cells were lysed on ice in a buffer containing 20 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, protease inhibitor and phosphatase inhibitor (Sigma-Aldrich). After 30 min of lysis, the cellular debris was removed by centrifugation at 13 200 r.p.m. for 15 min. Protein concentrations of lysates were determined by using the DC Protein Assay Kit (Bio-Rad, Hercules, CA, USA). The extracted proteins (10–30 μg) were separated by SDS-polyacrylamide gel electrophoresis and electrotransferred onto polyvinylidene difluoride membranes (GE Healthcare, Chalfont St Giles, Buckinghamshire, UK). Membranes were blocked with 5% skim milk, and proteins were detected by attaching the indicated primary antibodies overnight. Secondary antibodies conjugated with horseradish peroxidase were used for blotting and were visualized by a chemiluminescence reaction using ECL reagents (GE Healthcare). For cytoplasmic and nuclear fractionation, the cells were lysed on ice with 50 mM Tris-Cl, 2 mM EDTA, 0.1% NP-40, 10% glycerol and protease inhibitor. After 5 min of lysis, the samples were centrifuged for 5 min at 4000 revolution per min. Cytoplasmic supernatants were stored immediately, and nuclear pellets were further extracted in lysis buffer containing 20 mM Hepes–KOH, 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 10% glycerol, 25 mM β-glycerophosphate and protease inhibitor. After 10 min of lysis, nuclei were centrifuged at 15 000 revolutions per min and supernatant was collected. Nuclear (5 μg) and cytoplasmic (10 μg) fractions were subjected to western blot as described above.
Gene transduction and reporter assay
Genes for human RELA/P65 (NCBI Reference Sequence NM_021975.3) with a FLAG tag and human NFKBIA/IκBα (NCBI Reference Sequence NM_020529.2) with a FLAG tag were cloned into pcDNA3 (Invitrogen, Carlsbad, CA, USA). The plasmids (1–2 μg) were transiently transfected for 6 h using Genjet transfection reagent (SignaGen, Gaithersburg, MD, USA) according to the manufacturer’s instructions. For the reporter assay, four copies of the NF-κB consensus site (5′-GGGAATTTCC-3′) were cloned into the pGL3 luciferase reporter vector (Promega, San Luis Obispo, CA, USA). In addition, 4T1 or MDA-MB-231 (Supplementary Figure 2) cells were transiently transfected in six-well culture plates with NF-κB-luciferase reporter vector (1 μg) for 6 h. The cells were treated with CXCL10 or JN-2 for 24 h after transfection. Luciferase activity was measured using the dual-luciferase assay system (Promega) according to the manufacturer’s instructions.
Real-time PCR
Total cellular RNA was extracted by using TRIzol reagent (Invitrogen), and first-strand cDNAs were synthesized from 2 μg of RNA by using the SuperScript II Preamplification System (Invitrogen). Relative mRNA levels were determined using the ABI Prism 7500 sequence detection system with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) as previously described.25 Expression of target genes was determined according to the 2-ΔΔCT method using GAPDH as a reference gene. The following primer sequences were used: RANKL F: 5′-TGGAAGGCTCATGGTTGGAT-3′ and R: 5′-CATTGATGGTGAGGTGTGCA-3′; OPG F: 5′-TGGAACCCCAGAGCGAAACA-3′ and R: 5′-GCAGGAGGCCAAATGTGCTG-3′; CXCL10 F: 5′-ACTCCCCTTTACCCAGTGGA-3′ and R: 5′-GGACCATGGCTTGACCATCA-3′; GAPDH F: 5′-GAGAGTGTTTCCTCGTCCCG-3′ and R: 5′-ATGAAGGGGTCGTTGATGGC-3′.
Statistical analysis
All quantitative data are represented as the mean±s.d.’s (n⩾3). Two-group comparisons were analyzed with Student’s t-tests. P-values <0.05 were considered significant.
Results
The design of JN-2
We developed a new synthetic CXCR3 antagonist JN-2 for evaluation of its function on breast cancer cells. To develop JN-2, we synthesized benzoxazole derivatives and evaluated their biological activities as reported in our previous studies.26, 27, 28, 29 On the basis of the results of high-throughput screening30 from Abbot Laboratories that compound a (Figure 1) is effective as a CXCR3 antagonist, we designed our compound b, in which benzoxazole was expected to function as an isostere of benzimidazole. Compound b displayed effective CXCR3 antagonistic activity.31 Compound b was further modified to JN-2 (compound c), which contains an amide side chain to mimic the well-known CXCR3 antagonist AMG 487 (compound d), a small molecular antagonist that has been evaluated in Phase IIa clinical trials.32
JN-2 inhibits CXCL10 expression in 4T1 cells
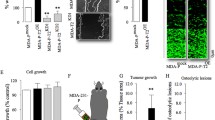
We first measured secreted CXCL10, a ligand for CXCR3, before investigation of the biological effect of JN-2 on 4T1 cells. Secreted CXCL10 levels were increased in 4T1 cells during the cell growth period (Figure 2a). We then investigated the effects of JN-2 on cell viability. Treatment with various doses of JN-2 for 24 h did not alter 4T1 cell viability (Figure 2b). We next questioned whether treatment with JN-2 would affect CXCL10 expression during a 24- h culture period. Interestingly, JN-2 suppressed CXCL10 mRNA expression in a dose-dependent manner (Figure 2c). Indeed, we observed that the well-known CXCR3 antagonist AMG 487 also reduced CXCL10 mRNA expression in 4T1 cells (Supplementary Figure 1). To confirm whether reduced CXCL10 mRNA expression affects its secretion level, we measured the amount of CXCL10 in the cultured supernatant of 4T1 cells. JN-2 suppressed the secretion level of CXCL10 during 12 and 24 h of culture (Figure 2d). We next investigated whether CXCL10 exerted an effect on endogenous mRNA expression in 4T1 cells. Stimulation of 4T1 cells with CXCL10 increased its mRNA expression (Figure 2e). We further observed that the increase in CXCL10 mRNA expression induced by CXCL10 stimulation was efficiently inhibited by JN-2 (Figure 2f). These data suggested that secretion of CXCL10 might contribute to endogenous CXCL10 expression through its receptor CXCR3.
Antagonism of CXCR3 inhibits CXCL10 expression in 4T1 cells. (a) 4T1 cells were cultured for the indicated times, and secreted CXCL10 levels were measured by ELISA (*P<0.05, **P<0.01). (b, c) 4T1 cells were cultured for 24 h with DMSO or various doses of JN-2. After culturing, cell viability (b) and CXCL10 mRNA levels (c) were analyzed by the EZ-Cytox assay kit and real-time PCR, respectively (*P<0.05). (d) 4T1 cells were cultured with DMSO or JN-2 (10 μM) for 12 or 24 h, and CXCL10 production was measured (*P<0.05, **P<0.01). (e) 4T1 cells were cultured with various doses of mCXCL10 for 24 h, CXCL10 mRNA levels were analyzed (*P<0.05). (f) 4T1 cells were cultured with mCXCL10 (300 ng ml−1) alone or together with JN-2 (10 μM) for 24 h, and CXCL10 mRNA levels were analyzed (*P<0.05).
Suppression of 4T1 motility by JN-2
CXCL10 and its receptor CXCR3 are crucial factors for migration and metastasis in various cancer cells.33 To investigate the inhibitory effect of the CXCR3 antagonist JN-2 on 4T1 cell motility, the wound healing assay was performed. Stimulation with CXCL10 increased 4T1 cell migration to the denuded zone after 24 h of incubation (Figure 3a). However, treatment with JN-2 suppressed CXCL10-induced migration and even the basal migration ability of 4T1 cells. To further examine the effect of the CXCR3 antagonist JN-2, we assessed the migratory potential of 4T1 cells using a transwell migration assay. Medium containing CXCL10 in the bottom of the migration chamber efficiently induced 4T1 cell migration at the upper chamber (Figure 3b). However, JN-2-containing medium in the bottom chamber inhibited 4T1 cell migration with or without CXCL10. These results suggest that 4T1 breast cancer cells require CXCR3 to gain their basal motility.
Antagonism of CXCR3 inhibits 4T1 motility. (a) Monolayers of confluent 4T1 cells were scratched. After culturing for 24 h with media containing DMSO or JN-2 (10 μM) and PBS or mCXCL10 (300 ng ml−1), cells that migrated to the denuded zone were analyzed (*P<0.05). Scale bar is 200 μm. (b) 4T1 cells in medium were placed in the upper chamber, and medium containing CXCL10 (300 ng ml−1) or JN-2 (10 μM) was placed in the bottom chamber. After 24 h of incubation, migrated cells were stained with H&E and analyzed (*P<0.05). Scale bar is 500 μm.
CXCL10/CXCR3 induces activation of NF-κB signaling
To address the mechanism by which 4T1 regulates CXCL10 expression accompanied by motility, we explored the effects of CXCL10 on the expression of the NF-κB subunit P65, one of the highly expressed transcription factors in cancer cells.34, 35 CXCL10 induced an upregulation of P65 in 4T1 cells after 24 h of stimulation, whereas the inhibitory subunit IκBα was downregulated (Figure 4a). We further investigated the effect of CXCL10 on the transcriptional activation of NF-κB activity by using a luciferase reporter gene under the κB promoter. CXCL10 stimulation induced the transcriptional activity of NF-κB (Figure 4b). To investigate whether blocking the CXCL10 receptor CXCR3 by its antagonists regulates the expression of P65, we cultured 4T1 cells in the presence of JN-2 or AMG 487. As expected, treatment with the CXCR3 antagonists for 24 h suppressed P65 expression but increased IκBα expression (Figure 4c). In addition, both basal and CXCL10-induced transcriptional activation of NF-κB were suppressed by JN-2 (Figure 4d). To investigate the effect of CXCL10 and JN-2 on the canonical NF-κB pathway, 4T1 cells were pre-treated with DMSO or JN-2 in serum-free medium for 1 h and then stimulated with CXCL10. DMSO-treated 4T1 cells exhibited increased P65 nuclear translocation within 30 min in response to CXCL10 stimulation (Figure 4e). However, we observed that pretreatment with JN-2 markedly reduced both basal and CXCL10-induced nuclear translocation of P65. This result suggests that CXCL10-induced activation of the NF-κB pathway requires CXCR3. To determine the role of p65 in CXCL10 expression, we transiently transfected Flag-tagged pcDNA3 or pcDNA3-P65 (P65) in 4T1 cells. Treatment with JN-2 for 24 h suppressed P65 expression (Figure 4f). However, forced expression of P65 abrogated the inhibitory effects of JN-2 on P65 expression. Next, we investigated whether P65 accounts for the inhibitory effect of JN-2 on CXCL10 expression. We found that the inhibitory effect of JN-2 on CXCL10 secretion was substantially blocked by forced expression of P65 (Figure 4g). These results indicated that CXCL10-induced P65 activation stimulates CXCL10 expression. In addition, we hypothesized that JN-2-induced IκBα expression may suppress NF-κB signaling activation and P65-induced CXCL10 expression. We then examined the role of IκBα in CXCL10 expression and NF-κB transcriptional activity. Transient transfection of IκBα inhibited expression of P65, CXCL10 secretion and NF-κB transcriptional activity in 4T1 cells (Figures 4h and i). These data suggest that CXCL10-induced P65 expression regulates CXCL10 secretion via canonical NF-κB transcriptional activation.
CXCL10 regulates the activation of canonical NF-κB signaling through CXCR3. (a) 4T1 cells were cultured with the indicated doses of mCXCL10 for 24 h. Total cell lysates were subjected to western blotting with the indicated antibodies. (b) 4T1 cells were transiently transfected with NF-κB reporter vector for 6 h. After incubation with PBS or mCXCL10 for 24 h, the luciferase assay was performed (**P<0.01). (c) 4T1 cells were cultured with the indicated doses of JN-2 or AMG 487 for 24 h. Total cell lysates were subjected to western blotting with the indicated antibodies. (d) 4T1 cells were transfected with NF-κB reporter vector for 6 h. After incubation with DMSO or JN-2 (10 μM) for 24 h, luciferase assay was performed (*P<0.05). (e) 4T1 cells were cultured with DMSO or JN-2 (10 μM) for 1 h in serum-free medium and treated with mCXCL10 (300 ng ml−1) for the indicated times. After nuclear and cytosolic fractionation, lysates were subjected to western blotting with the indicated antibodies. (f, g) 4T1 cells were transiently transfected with pcDNA3-Flag-empty or pcDNA3-Flag-P65 for 6 h. Transduced cells were further incubated with DMSO or JN-2 (10 μM) for 24 h. After culturing, cell lysates and CXCL10 protein secretion were analyzed by western blotting with the indicated antibodies (f) and ELISA (g; *P<0.05). (h) 4T1 cells were transfected with pcDNA3-Flag-empty (Veh) or pcDNA3-Flag-IκBα (Flag-IκBα) for 6 h. After incubation with fresh media for 24 h, cell lysates and CXCL10 protein secretion were analyzed by western blotting with the indicated antibodies (left) and ELISA (right; *P<0.05). (i) 4T1 cells were co-transfected with NF-κB reporter vector with pcDNA3-Flag-empty (Veh) or pcDNA3-Flag-IκBα (Flag-IκBα) for 6 h. After incubation with fresh media for 24 h, the luciferase assay was performed (**P<0.01).
Administration of JN-2 prevents 4T1-mediated bone destruction
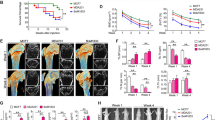
CXCR3 expression in cancer cells is implicated in osteolytic bone metastasis.20 We therefore examined the effects of the CXCR3 antagonist JN-2 on osteolytic bone metastasis of breast cancer. We performed an intrafemoral injection of 4T1 in female BALB/c mice (n=7 per group) to generate bone metastases. H&E staining and μCT revealed severe destruction of bone in 4T1-injected femur with DMSO compared with control mice (Figure 5a). Moreover, DMSO-injected mice exhibited extensive tumor spreading even on the outer surface of bone. However, injection of JN-2 inhibited the extensive 4T1-induced destruction of bone. We observed that 4T1-induced destruction of cortical and trabecular bone was inhibited by JN-2 compared with DMSO-injected mice (Figure 5b). Consistent with this finding, serum PYD levels, a marker of bone resorption, were increased by 4T1 cells, which were significantly inhibited by JN-2 treatment (Figure 5c). Given that JN-2 reduced P65 expression in vitro, we further assessed P65 expression in the bone area in breast cancer cells. We observed complex interactions between 4T1 cells and bone marrow cells with high P65 expression (brown) in the DMSO-injected mice (Figure 5d, top). In addition, we found that tumors in the outer surface of cortical bone exhibited reduced P65 expression compared with tumors in the bone marrow. In the case of JN-2-injected mice, we also observed P65 expression in the tumor area; however, the region of tumor and marrow was divided with lower P65 expression in the bone marrow area compared with DMSO-injected mice (Figure 5d, bottom). To address whether the inhibitory effect of JN-2 on bone destruction was caused by inhibition of osteoclast formation, we analyzed osteoclast number at the tumor-bone interface. We observed a reduced number of osteoclasts in JN-2-treated mice compared with the DMSO-treated mice (Figure 5e). We next investigated CXCL10 and RANKL expression in bone tissues and observed the inhibitory effect of JN-2 on 4T1-induced upregulation of CXCL10 and RANKL expression (Figure 5f).
JN-2 inhibits 4T1-induced bone destruction. Six-week-old female BALB/c (n=7) mice were intraperitoneally injected with DMSO or JN-2 (10 mg kg−1 of body weight) every other day before 4T1 injection. After 7 days, 4T1 cells (1 × 104 cells in 5 μl PBS) were injected into the femoral marrow space through the femoral condyle with a 30-gauge Hamilton syringe. Then, DMSO or JN-2 was intraperitoneally injected every other day for 21 days. (a) The femurs were investigated by histological sectioning with H&E staining (top) and μCT analysis (bottom and right; *P<0.05, **P<0.01). (b) The femurs were further analyzed for cortical bone volume (left) and trabecular number (right). *P<0.05, **P<0.01. Scale bar is 1 mm. (c) Serum PYD levels were measured by ELISA (*P<0.05, **P<0.01). (d) Histological sections of the femurs were immunostained with anti-P65 antibody (brown). T, tumor; M, marrow. Scale bar is 500 μm (left) and 100 μm (right). (e) Histological sections were stained for TRAP activity to detect osteoclast cells, and TRAP-positive cells were analyzed (*P<0.05). T, tumor; B, bone. Scale bar is 200 μm. (f) Total RNA were extracted from the femurs, and CXCL10 and RANKL mRNA levels were analyzed by real-time PCR (*P<0.05).
JN-2 inhibits osteoclast differentiation indirectly
Our findings of the inhibitory effects of JN-2 on tumor outgrowth and osteoclast formation in bone led us to investigate the mechanism underlying the inhibitory action of JN-2 on osteoclast formation. We previously reported that CXCL10 does not affect RANKL-induced osteoclast differentiation from its precursors.20 Consistent with this finding, JN-2 did not affect RANKL-induced osteoclast differentiation in BMM cultures (data not shown). CM obtained from 4T1 cell cultures exhibited increased RANKL-induced osteoclast differentiation in BMMs, which was not affected by JN-2 (Figure 6a). These results raised the possibility that JN-2 might inhibit osteoclast differentiation indirectly by affecting the ability of osteoblast lineage cells to stimulate osteoclast differentiation. Thus, we next examined the expression of RANKL and its decoy receptor OPG in osteoblasts. CXCL10 induced RANKL expression in osteoblasts; however, JN-2 did not affect RANKL or OPG expression (Figure 6b). These results led us to investigate whether CM from JN-2-treated 4T1 cells would inhibit osteoclast formation in co-culture of osteoblasts and osteoclast precursor cell BMMs. Interestingly, CM from DMSO-treated 4T1 cells efficiently formed osteoclasts, whereas CM from JN-2-treated 4T1 cells substantially suppressed osteoclast formation in the co-culture system (Figure 6c). Next, we examined whether CM from JN-2-treated 4T1 cells inhibited RANKL expression. CM from DMSO-treated 4T1 cells stimulated RANKL expression in osteoblasts, and CM from JN-2 (10 and 20 μM)-treated 4T1 cells reduced RANKL expression compared with CM from DMSO-treated 4T1 cells (Figure 6d). These results suggest that JN-2 affects osteoclast formation indirectly by inhibiting the ability of 4T1 cells to stimulate RANKL expression in osteoblasts. In addition, this discovery raises the possibility that antagonism of CXCR10/CXCR3 in cancer may have the clinical potential to regulate cancer-induced RANKL induction.36
JN-2 inhibits osteoclast differentiation indirectly. (a) BMMs were cultured in the presence of M-CSF (60 ng ml−1) and RANKL (RL; 100 ng ml−1) with or without CM+DMSO or CM+JN-2 (10 μM) for 4 days. Differentiated cells were stained for TRAP, and cells containing 3 or more nuclei were counted. Scale bar is 100 μm. (b) Mouse-calvarial osteoblasts were cultured with DMSO or JN-2 (10 μM) in the presence of PBS or CXCL10 (300 ng ml−1). After 24 h of incubation, RANKL and OPG mRNA levels were analyzed by real-time PCR. (c) BMMs and mouse-calvarial osteoblasts were co-cultured for 9 days with or without CM from 4T1 cells treated with DMSO or JN-2 (10 μM). The cells were stained for TRAP, and cells containing three or more nuclei were counted. Scale bar is 100 μm (*P<0.05). (d) Mouse-calvarial osteoblasts were cultured for 24 h with or without CM from 4T1 cells treated with DMSO or JN-2. RANKL and OPG mRNA levels were analyzed by real-time PCR (*P<0.05).
Discussion
Recently, increasing evidence suggests a crucial role of CXCR3 in metastasis, and inhibition of CXCR3 has emerged as a therapeutic target in cancer.37, 38, 39, 40, 41, 42 Although numerous studies have demonstrated that blocking CXCR3 has an anti-metastatic effect on tumor cells, in the present study, we focused on CXCL10/CXCR3-mediated intracellular signaling and metastasis-induced bone destruction. We observed that the murine breast cancer cell line 4T1 secreted the CXCR3 ligand CXCL10; however, CXCL10 mRNA expression was inhibited by treatment with the CXCR3 antagonists JN-2 and AMG 487 without CXCL10 stimulation. Moreover, the migration ability of 4T1 cells was inhibited by JN-2 regardless of CXCL10 stimulation. These results suggest that highly expressed CXCL10 may contribute to sustain tumor progression through CXCR3 via cell-autonomous regulation. Insight into the physiological actions of CXCL10 was provided by data demonstrating that under the influence of IFN-γ, the host immune response induces an amplification feedback loop of CXCL10 secretion by several cell types, including Th1 lymphocytes, endothelial cells, fibroblasts and keratinocytes.43 In addition, we previously showed that cancer cells augment CXCL10 production from macrophages in a cell–cell contact manner and that host-deficiency of CXCL10 inhibits osteolytic bone metastasis.20 Thus, together, these findings suggest that autocrine and paracrine production of CXCL10 facilitates cell motility and osteolytic bone metastasis of cancer cells through activation of CXCR3 in cancer cells.
STAT-1, NF-kB, and the transcriptional coactivator CREB-binding protein are involved in the transcriptional activation of CXCL10.44 NF-κB has been elucidated as a regulator of the inflammatory response and immune cell function. However, it is now apparent that activation of NF-κB in cancer is common. For instance, oncoproteins, such as Bcr-Abl and Ras, require NF-κB signaling activation.45 In this study, we found that CXCL10 increases the expression of the NF-κB subunit P65 and NF-κB transcriptional activity in 4T1 cells. We also observed that CXCL10 induces NF-κB transcriptional activity in the human breast cancer cell line MDA-MB-231 (Supplementary Figure 2). In addition, CXCL10 rapidly induced nuclear translocation of P65 within 30 min in 4T1 cells. In contrast, the CXCR3 antagonists AMG 487 and JN-2 inhibited the expression and nuclear translocation of P65 regardless of CXCL10 stimulation. Forced expression of P65 abrogated the inhibitory effects of JN-2 on P65 expression and CXCL10 secretion. By contrast, overexpression of IκBα reduced P65 expression, secreted CXCL10 and NF-κB transcriptional activity. Thus, these results suggest that the CXCL10/CXCR3 axis creates a positive feedback loop through activation of the canonical NF-κB signaling pathway, resulting in the maintenance of high NF-κB activity in 4T1 cells.
Chemokine receptors have been the target of drug discovery with a focus on the development of a new drug for small molecule antagonists that are capable of blocking the action of chemokine-induced receptor activation. The selective CXCR3 antagonist SCH 546738 attenuates the development of autoimmune diseases.46 The CXCR3 antagonist AMG 487 exhibits inhibitory effects on lung metastasis in breast cancer.47 In the present study, we demonstrated that JN-2 inhibits osteolytic bone metastasis of 4T1 cells with a decrease in tumor outgrowth, P65 expression, osteoclast formation, and the expression of RANKL and CXCL10. Our in vitro experiments demonstrated that JN-2 does not affect 4T1 CM-induced osteoclast differentiation but inhibits osteoclast differentiation indirectly by suppression of the ability of 4T1 cells to stimulate RANKL expression in osteoblasts. We previously demonstrated that inhibition of CXCR3 by gene knockdown in cancer cells reduces osteolytic bone metastasis.20 Thus, our in vitro and in vivo results suggest that the direct inhibitory action of JN-2 on CXCR3 in 4T1 cells contributes to its inhibitory effects on osteolytic bone metastasis of 4T1 cells. However, we could not exclude the possibility of the influence of JN-2 on host cells. Indeed, we observed that JN-2 inhibits 4T1-induced upregulation of P65 expression in the bone marrow region. Thus, whether specific inhibition of CXCR3 in host cells also affects osteolytic bone metastasis should be assessed in further studies.
The factors secreted from breast cancer cells, including IL-1, IL-6, prostaglandin E2 (PGE2) and TNFα, play a crucial role in breast cancer-mediated osteoclast activation.48, 49 These secreted factors stimulate osteoblasts to produce RANKL.2, 36 IL-1 and IL-6 stimulate RANKL expression via activation of the gp130-STAT3 signaling axis in stromal/osteoblastic cells.50 PGE2 is a stimulator of bone resorption, and the major effect of PGE2 on resorption is considered to occur indirectly through EP4 receptor-mediated RANKL expression in osteoblastic cells.51 In addition, IL-1 and TNFα induce RANKL expression via P38 MAPK signaling.52 We previously found that CXCL10 increases RANKL mRNA and protein expression in osteoblasts through Toll-like receptor 4 but not CXCR3.20 Consistent with this finding, treatment of osteoblasts with JN-2 did not affect CXCL10-induced RANKL expression in osteoblasts. We also found that JN-2 suppresses CXCL10 secretion from 4T1 cells and that CM from 4T1 cells treated with JN-2 reduces RANKL expression in osteoblasts compared with control 4T1 CM. These findings indicate that CXCL10 is one of the factors secreted from 4T1 cells that stimulates RANKL expression in osteoblasts. Our results also suggest that JN-2-induced reduced CXCL10 secretion from 4T1 cells contributes, at least in part, to its inhibitory effect on RANKL expression in osteoblasts and osteoclast differentiation. However, whether the CXCL10/CXCR3/NF-kB feedback loop also affects other secreted factors from 4T1 cells that stimulate RANKL expression in osteoblasts remains unclear.
In conclusion, this study shows that the CXCL10/CXCR3 axis in breast cancer 4T1 cells creates a positive feedback loop through activation of the canonical NF-κB signaling pathway, which stimulates cell motility and osteolytic bone metastasis of 4T1 cells. Therefore, we propose that the CXCL10/CXCR3/NF-κB signaling pathway plays a fundamental role in malignant tumor properties.
References
Akhtari M, Mansuri J, Newman KA, Guise TM, Seth P . Biology of breast cancer bone metastasis. Cancer Biol Ther 2008; 7: 3–9.
Chen YC, Sosnoski DM, Mastro AM . Breast cancer metastasis to the bone: mechanisms of bone loss. Breast Cancer Res 2010; 12: 215.
Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 2003; 3: 537–549.
Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R et al. TGF-beta signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest 1999; 103: 197–206.
Mundy GR . Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer 2002; 2: 584–593.
Johrer K, Pleyer L, Olivier A, Maizner E, Zelle-Rieser C, Greil R . Tumour-immune cell interactions modulated by chemokines. Expert Opin Biol Ther 2008; 8: 269–290.
Wang JM, Deng X, Gong W, Su S . Chemokines and their role in tumor growth and metastasis. J Immunol Methods 1998; 220: 1–17.
Raman D, Baugher PJ, Thu YM, Richmond A . Role of chemokines in tumor growth. Cancer Lett 2007; 256: 137–165.
Baggiolini M . Chemokines and leukocyte traffic. Nature 1998; 392: 565–568.
Gillitzer R, Goebeler M . Chemokines in cutaneous wound healing. J Leukoc Biol 2001; 69: 513–521.
Balkwill F, Mantovani A . Inflammation and cancer: back to Virchow? Lancet 2001; 357: 539–545.
Mantovani A, Savino B, Locati M, Zammataro L, Allavena P, Bonecchi R . The chemokine system in cancer biology and therapy. Cytokine Growth Factor Rev 2010; 21: 27–39.
Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME et al. Involvement of chemokine receptors in breast cancer metastasis. Nature 2001; 410: 50–56.
Liang Z, Yoon Y, Votaw J, Goodman MM, Williams L, Shim H . Silencing of CXCR4 blocks breast cancer metastasis. Cancer Res 2005; 65: 967–971.
Balkwill F . Cancer and the chemokine network. Nat Rev Cancer 2004; 4: 540–550.
Rubie C, Kollmar O, Frick VO, Wagner M, Brittner B, Graber S et al. Differential CXC receptor expression in colorectal carcinomas. Scand J Immunol 2008; 68: 635–644.
Sun RH, Wang GB, Li J, Cui J . [Role of CCL21/CCR7 in invasion of colorectal carcinoma cell line SW480]. Ai Zheng 2009; 28: 708–713.
Gangur V, Birmingham NP, Thanesvorakul S . Chemokines in health and disease. Vet Immunol Immunopathol 2002; 86: 127–136.
Luster AD, Ravetch JV . Biochemical characterization of a gamma interferon-inducible cytokine (IP-10). J Exp Med 1987; 166: 1084–1097.
Lee JH, Kim HN, Kim KO, Jin WJ, Lee S, Kim HH et al. CXCL10 promotes osteolytic bone metastasis by enhancing cancer outgrowth and osteoclastogenesis. Cancer Res 2012; 72: 3175–3186.
Harris DP, Bandyopadhyay S, Maxwell TJ, Willard B, DiCorleto PE . Tumor necrosis factor (TNF)-alpha induction of CXCL10 in endothelial cells requires protein arginine methyltransferase 5 (PRMT5)-mediated nuclear factor (NF)-kappaB p65 methylation. J Biol Chem 2014; 289: 15328–15339.
Shen Q, Zhang R, Bhat NR . MAP kinase regulation of IP10/CXCL10 chemokine gene expression in microglial cells. Brain Res 2006; 1086: 9–16.
Majumder S, Zhou LZ, Chaturvedi P, Babcock G, Aras S, Ransohoff RM . p48/STAT-1alpha-containing complexes play a predominant role in induction of IFN-gamma-inducible protein, 10 kDa (IP-10) by IFN-gamma alone or in synergy with TNF-alpha. J Immunol 1998; 161: 4736–4744.
Kim SD, Kim HN, Lee JH, Jin WJ, Hwang SJ, Kim HH et al. Trapidil, a platelet-derived growth factor antagonist, inhibits osteoclastogenesis by down-regulating NFATc1 and suppresses bone loss in mice. Biochem Pharmacol 2013; 86: 782–790.
Lee JH, Kim HN, Yang D, Jung K, Kim HM, Kim HH et al. Trolox prevents osteoclastogenesis by suppressing RANKL expression and signaling. J Biol Chem 2009; 284: 13725–13734.
Yoon JH, Song H, Kim SW, Han G, Choo HYP . A facile synthesis of 2-aminothiazolo[5,4-b]pyridines and 2-aminobenzoxazoles via cyclization of thioureas. Heterocycles 2005; 65: 2729–2740.
Lee JH, An MH, Choi EH, Choo HYP, Han G . A facile synthesis of 2-acyl and 2-alkylaminobenzimidazoles for 5-lipoxygenase inhibitors. Heterocycles 2006; 70: 571.
Kim BJ, Kim J, Kim YK, Choi SY, Choo HYP . Synthesis of benzoxazole amides as novel antifungal agents against Malassezia Furfur. Bull Korean Chem Soc 2010; 31: 1270–1274.
Song H, Oh SR, Lee HK, Han G, Kim JH, Chang HW et al. Synthesis and evaluation of benzoxazole derivatives as 5-lipoxygenase inhibitors. Bioorg Med Chem 2010; 18: 7580–7585.
Hayes ME, Wallace GA, Grongsaard P, Bischoff A, George DM, Miao WY et al. Discovery of small molecule benzimidazole antagonists of the chemokine receptor CXCR3. Bioorg Med Chem Lett 2008; 18: 1573–1576.
Lee ZH, Choo HYP Preparation of benzoxazole derivatives as CXCR3/CXCL10 antagonists. Korea Patent 1020130039801. 11 April 2016.
Andrews SP, Cox RJ . Small molecule CXCR3 Antagonists. J Med Chem 2016; 59: 2894–2917.
Liu M, Guo S, Stiles JK . The emerging role of CXCL10 in cancer (review). Oncol Lett 2011; 2: 583–589.
Karin M, Cao Y, Greten FR, Li ZW . NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer 2002; 2: 301–310.
Orlowski RZ, Baldwin AS Jr . NF-kappaB as a therapeutic target in cancer. Trends Mol Med 2002; 8: 385–389.
Martin TJ . Bone biology and anabolic therapies for bone: current status and future prospects. J Bone Metab 2014; 21: 8–20.
Kawada K, Sonoshita M, Sakashita H, Takabayashi A, Yamaoka Y, Manabe T et al. Pivotal role of CXCR3 in melanoma cell metastasis to lymph nodes. Cancer Res 2004; 64: 4010–4017.
Kawada K, Hosogi H, Sonoshita M, Sakashita H, Manabe T, Shimahara Y et al. Chemokine receptor CXCR3 promotes colon cancer metastasis to lymph nodes. Oncogene 2007; 26: 4679–4688.
Cambien B, Karimdjee BF, Richard-Fiardo P, Bziouech H, Barthel R, Millet MA et al. Organ-specific inhibition of metastatic colon carcinoma by CXCR3 antagonism. Br J Cancer 2009; 100: 1755–1764.
Zhu GQ, Yan HH, Pang YL, Jian J, Achyut BR, Liang XH et al. CXCR3 as a molecular target in breast cancer metastasis: inhibition of tumor cell migration and promotion of host anti-tumor immunity. Oncotarget 2015; 6: 43408–43419.
Walser TC, Rifat S, Ma XR, Kundu N, Ward C, Goloubeva O et al. Antagonism of CXCR3 inhibits lung metastasis in a murine model of metastatic breast cancer. Cancer Res 2006; 66: 7701–7707.
Pradelli E, Karimdjee-Soilihi B, Michiels JF, Ricci JE, Millet MA, Vandenbos F et al. Antagonism of chemokine receptor CXCR3 inhibits osteosarcoma metastasis to lungs. Int J Cancer 2009; 125: 2586–2594.
Antonelli A, Ferrari SM, Giuggioli D, Ferrannini E, Ferri C, Fallahi P . Chemokine (C-X-C motif) ligand (CXCL)10 in autoimmune diseases. Autoimmun Rev 2014; 13: 272–280.
Clarke DL, Clifford RL, Jindarat S, Proud D, Pang L, Belvisi M et al. TNFalpha and IFNgamma synergistically enhance transcriptional activation of CXCL10 in human airway smooth muscle cells via STAT-1, NF-kappaB, and the transcriptional coactivator CREB-binding protein. J Biol Chem 2010; 285: 29101–29110.
Baldwin AS . Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest 2001; 107: 241–246.
Jenh CH, Cox MA, Cui L, Reich EP, Sullivan L, Chen SC et al. A selective and potent CXCR3 antagonist SCH 546738 attenuates the development of autoimmune diseases and delays graft rejection. BMC Immunol 2012; 13: 2.
Walser TC, Rifat S, Ma X, Kundu N, Ward C, Goloubeva O et al. Antagonism of CXCR3 inhibits lung metastasis in a murine model of metastatic breast cancer. Cancer Res 2006; 66: 7701–7707.
Roodman GD . Mechanisms of bone metastasis. N Engl J Med 2004; 350: 1655–1664.
Esquivel-Velazquez M, Ostoa-Saloma P, Palacios-Arreola MI, Nava-Castro KE, Castro JI, Morales-Montor J . The role of cytokines in breast cancer development and progression. J Interferon Cytokine Res 2015; 35: 1–16.
O'Brien CA, Gubrij I, Lin SC, Saylors RL, Manolagas SC . STAT3 activation in stromal/osteoblastic cells is required for induction of the receptor activator of NF-kappaB ligand and stimulation of osteoclastogenesis by gp130-utilizing cytokines or interleukin-1 but not 1,25-dihydroxyvitamin D3 or parathyroid hormone. J Biol Chem 1999; 274: 19301–19308.
Suzawa T, Miyaura C, Inada M, Maruyama T, Sugimoto Y, Ushikubi F et al. The role of prostaglandin E receptor subtypes (EP1, EP2, EP3, and EP4) in bone resorption: an analysis using specific agonists for the respective EPs. Endocrinology 2000; 141: 1554–1559.
Rossa C, Ehmann K, Liu M, Patil C, Kirkwood KL . MKK3/6-p38 MAPK signaling is required for IL-1beta and TNF-alpha-induced RANKL expression in bone marrow stromal cells. J Interferon Cytokine Res 2006; 26: 719–729.
Acknowledgements
This study was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science, ICT & Future Planning (NRF-2014R1A2A2A01002531) and a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare (A120850).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Jin, W., Kim, B., Kim, D. et al. NF-κB signaling regulates cell-autonomous regulation of CXCL10 in breast cancer 4T1 cells. Exp Mol Med 49, e295 (2017). https://doi.org/10.1038/emm.2016.148
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/emm.2016.148
- Springer Nature Limited
This article is cited by
-
A review of the pleiotropic actions of the IFN-inducible CXC chemokine receptor 3 ligands in the synovial microenvironment
Cellular and Molecular Life Sciences (2023)
-
CHMP2A regulates tumor sensitivity to natural killer cell-mediated cytotoxicity
Nature Communications (2022)
-
Rg1 exerts protective effect in CPZ-induced demyelination mouse model via inhibiting CXCL10-mediated glial response
Acta Pharmacologica Sinica (2022)
-
Systemic CXCL10 is a predictive biomarker of vitiligo lesional skin infiltration, PUVA, NB-UVB and corticosteroid treatment response and outcome
Archives of Dermatological Research (2022)
-
DACH1 inhibits breast cancer cell invasion and metastasis by down-regulating the transcription of matrix metalloproteinase 9
Cell Death Discovery (2021)