Abstract
We report the neuroprotective role of FAS apoptotic inhibitory molecule 2 (FAIM2), an inhibitor of the FAS signaling pathway, during stress-induced photoreceptor apoptosis. Retinal detachment resulted in increased FAIM2 levels in photoreceptors with higher amounts detected at the tips of outer segments. Activation of FAS death receptor via FAS-ligand led to JNK-mediated FAIM2 phosphorylation, decreased proteasome-mediated degradation and increased association with the FAS receptor. Photoreceptor apoptosis was accelerated in Faim2 knockout mice following experimental retinal detachment. We show that FAIM2 is primarily involved in reducing stress-induced photoreceptor cell death but this effect was transient. FAIM2 was found to interact with both p53 and HSP90 following the activation of the FAS death pathway and FAIM2/HSP90 interaction was dependent on the phosphorylation of FAIM2. Lack of FAIM2 led to increased expression of proadeath genes Fas and Ripk1 in the retina under physiologic conditions. These results demonstrate that FAIM2 is an intrinsic neuroprotective factor activated by stress in photoreceptors and delays FAS-mediated photoreceptor apoptosis. Modulation of this pathway to increase FAIM2 expression may be a potential therapeutic option to prevent photoreceptor death.
Similar content being viewed by others
Main
Separation of outer retina from the retinal pigment epithelium (RPE) is a common form of injury that may occur alone in retinal detachment or with other pathologic processes such as age-related macular degeneration or diabetic retinopathy. Despite significant advances in the medical and surgical management of retina-RPE separation, patients often lose vision, primarily due to the death of photoreceptors.1, 2 Our previous studies demonstrated that the main pathologic event causing photoreceptor death following photoreceptor-RPE separation is the activation of the apoptotic FAS signaling pathway.3, 4, 5 Preventing FAS pathway activity provides significant protection against separation-induced death of the photoreceptors.5, 6
Despite rapid activation of apoptosis after retina-RPE separation, a significant number of photoreceptors survive for extended periods of time.4, 5 The clinical correlation of this observation is that patients with retinal detachments affecting the macula can recover significant vision after repair, especially if repaired within one week.7, 8 These experimental and clinical observations suggest that anti-apoptotic pathways are activated within the retina to counteract the effect of pro-apoptotic signals. We have shown the significance of several such pathways in the retina, including interleukin-6 (IL6) signaling and autophagy.6, 9
Previous gene microarray analysis of experimental detachments revealed increased expression of genes involved in FAS-receptor signaling and stress-response pathways, including Faim2, coding for FAS apoptotic inhibitory molecule 2 (FAIM2).10 FAIM2 is an evolutionarily conserved protein and is predominantly expressed in neuronal cells as a 35 kDa membrane protein.11 FAIM2 was shown to prevent apoptosis by direct interaction with FAS upstream of FAS-associated death domain containing protein (FADD).12 FAIM2 expression in cerebellar granule neurons increases their resistance to FAS mediated apoptosis.13 Neurons of Faim2-deficient mice are more susceptible to combined oxygen–glucose deprivation in vitro and caspase-associated cell death and neurological impairment after cerebral ischemia in vivo.14 Similarly, FAIM2 is required for the development and survival of granular and Purkinje cells.15
We have previously demonstrated that retinal detachment results in increased FAIM2 protein levels in vivo, and this finding is reproduced in vitro by exogenous activation of FAS signaling in 661 W cells.16 We now report evidence demonstrating the neuroprotective role of FAIM2 in the retina following photoreceptor stress. To better characterize the regulation and function of FAIM2, we investigated the location of FAIM2 expression in the retina and mechanism of FAIM2 upregulation following FAS activation, including identification of proteins interacting with FAIM2 and the effect of FAIM2 on the transcription of pro- and anti-apoptotic genes known to be critical for photoreceptor death.
Results
Retinal detachment leads to an increase in FAIM2 levels in photoreceptors
We previously reported increased FAIM2 protein levels in the experimental retinal detachment model.16 To determine the location of FAIM2 in detached retinas, we performed immunohistochemistry in mice 24 h after experimental photoreceptor-RPE separation. FAIM2 levels primarily increased in the outer nuclear layer of the detached retinas, consistent with elevated levels in the detached photoreceptors (Figure 1a). Although the FAIM2 signal was seen throughout the outer nuclear layer (ONL) and inner segment/outer segment (IS/OS) layers after retinal detachment, most of the staining was concentrated at the OS of photoreceptors.
Retinal detachment leads to increased FAIM2 levels in the photoreceptors. Whole-mouse retinas were harvested following detachments at 24 h. (a) Immunohistochemistry shows increased levels of FAIM2 in the detached retina in the outer nuclear layer. Magnified view on the right demonstrates FAIM2 staining preferentially in the outer segments of photoreceptors. GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer; IS/OS, Inner segment/Outer segment; RPE, Retinal pigment epithelium, N=3. (b) FAIM2 immunoreactivity on western blot following 24-h retinal detachment in mice. Increased FAIM2 protein in detected after retinal detachment. Actin serves as the loading control. Normalization of FAIM2 levels to actin levels shows 3-fold increase of FAIM2 expression in detached retinas (mean±S.E.M., N=4, P<0.05). (c) Increase in FAIM2 levels after 24 h of retinal detachment is independent of Faim2 gene transcription. Quantitative RT-PCR analysis of Faim2 with β-actin mRNA serving as control. Relative expression was analyzed by comparative threshold cycle (2−ΔΔct) method. Expression values were represented as fold change over control after normalization with actin. Data represent mean±S.E.M., N=3
We performed western blot analysis to quantitate the amount of FAIM2 upregulation following retinal detachment (Figure 1b). We found that retinal detachment led to approximately 3-fold increase in the amount of FAIM2 protein at 24 h. Although the level of FAIM2 protein increases with retinal detachment by 24 h (Figures 1a and b), RT-PCR did not detect any increase in Faim2 transcript levels in the detached retinas (Figures 1c). Our data strongly suggests that the increased level of FAIM2 found immediately after retinal detachment is the result of increased stability of FAIM2 protein, rather than increased gene transcription.
FAIM2 mediates photoreceptor neuroprotection after retinal detachment
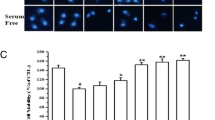
Our previous work showed that FAIM2 regulated caspase 8 activation and reduced apoptosis in 661 W cells following exogenous activation of FAS receptor signaling.16 The 661 W cell is a murine cone-derived cell line,17 and has been used extensively in the study of photoreceptor cell biology. To confirm the role of FAIM2 in retinal detachment induced photoreceptor apoptosis, experimental retinal detachments were created in wild-type and Faim2−/− mice. In rodent eyes, the peak of TUNEL staining occurs at 3 days after retina-RPE separation, with a rapid decline in TUNEL-positive cells to near pre-separation levels by day 7.6 As expected, wild-type mice showed a peak of TUNEL-positive staining of the photoreceptors at 3 days after creation of the retinal detachment (Figures 2a and b). In Faim2 knockout animals, TUNEL staining followed the same time course, with peak of TUNEL-positive staining at 3 days (Figure 2a). As predicted by our hypothesis, the level of TUNEL-positive staining was much more robust with a near doubling of the number of dying cells in knockout retinas (Figure 2b). Although the knockout animals had increased TUNEL staining in the ONL in all tested time points, the difference was only statistically significant at 3 days when the apoptotic activity is the highest.
Lack of FAIM2 leads to increased apoptosis in stressed photoreceptors. (a) Immunofluorescence staining of TUNEL (green) and DAPI (blue) in the outer nuclear layer (ONL), N=3 (b) Quantification of TUNEL-positive cells in the ONL. Significant increased photoreceptor death observed at 3-day detachment in the Faim2−/− mice compared to wild-type animals. Mean±S.E.M.; n=8–12, P<0.05, N=3
To determine if FAIM2 is important for long-term survival of photoreceptors, experimental retinal detachments were induced in wild type and Faim2−/− mice and maintained for 1 and 2 months. Photoreceptor cell counts were assessed by the number of photoreceptor nuclei in the ONL and normalized to the total retinal area. The number of photoreceptors was similar between wild-type mice and Faim2−/− mice at baseline (Figures 3a and b). There was a decline in the normalized ONL cell count at the 1 month time point for Faim2−/− mice, in contrast to wild-type animals, which showed no detectable cell loss (P=0.0014; Figures 3a and b). Similarly, ONL area showed a decline in the Faim2 knockout retinas by 1 month after detachment while there was little effect seen in wild-type controls. Two months after detachment, there was further decline in the ONL cell count and ONL area in both experimental groups. Both wild-type and Faim2−/− animals showed similar levels of remaining cells in the ONL at 2 months, demonstrating that FAIM2 is important for delaying photoreceptor apoptosis but is unable to prevent cell death during periods of prolonged detachment (Figure 3).
The absence of FAIM2 leads to increased photoreceptor loss in the ONL following retinal detachment. Retinal cross sections from the eyes of wild-type (Faim2+/+) and Faim2 knockout (Faim2−/−) mice were stained with toluidine blue 1-month and 2-month post detachment. (a) Retinal section of attached and detached retina, 1 month after creation of detachment. (b) Retinal section of attached and detached retina, 2 month after creation of detachment. (c) Graph summarizing photoreceptor nuclear counts and ONL area between the attached and detached retina after 1-month detachment. Cell death in the detached retina was significantly higher in Faim2−/− mice compared to Faim2+/+ mice based on nuclear counts and ONL area measurements. Mean±S.E.M., n=8–11, P=0.0014. (d) There was no statistically significant difference in cell death 2 months after detachment in wild type or knockout animals (n=8–11)
Faim2 expression is not required for normal development or postnatal survival of photoreceptors
Our data demonstrates the importance of FAIM2 in preventing stress-induced photoreceptor apoptosis. We next investigated the role of FAIM2 in photoreceptor survival with age. Genetic deletion of Faim2 had no discernable effect on retinal anatomy and all retinal structures were anatomically normal in wild-type and knockout mice aged to 6 and 12 months (Figure 4a). Assessment of nuclear numbers in the ONL showed that the number of photoreceptors were similar in wild-type and Faim2 knockout animals (Figure 4b). Similarly, ONL area measurements confirmed that lack of FAIM2 is not necessary for photoreceptor survival under physiologic conditions (Figure 4b). These results clearly demonstrate that FAIM2 function is not necessary for photoreceptor development and survival under physiologic conditions or with aging.
FAIM2 is not required for the survival of photoreceptors with age. (a) Eyes were enucleated at 6 month and 1 year and retinal sections were stained with 0.5% toluidine blue for histology. (b) ONL cell counts and ONL area normalized to retinal area. No significant changes in the number of cells in the ONL or changes in morphology were observed for Faim2+/+ or Faim2−/− mice. Mean±S.E.M., n=10. GCL, ganglion cell layer; INL, inner nuclear layer; IS/OS, inner segment/outer segment; ONL, outer nuclear layer; RPE, retinal pigment epithelium
FAIM2 binds FAS during photoreceptor apoptosis
FAIM2 was initially identified as an inhibitor of the FAS death receptor-signaling pathway.12 To determine whether FAIM2 interacts directly with FAS in the retina, we immunoprecipitated FAS and FAIM2. FAS–FAIM2 interaction was seen early during retinal detachment and peaked at 12 h after retina-RPE separation (Figures 5a and b). Both FAIM2 and FAS protein levels remained unchanged in the first 12 h of retinal detachment, indicating that increased association of FAIM2 with FAS is mostly due to factors other than increased protein concentration (Figure 5a). There was reduced FAIM2–FAS interaction by 24 h, indicating that early post-detachment signaling results in activation of FAIM2, and that FAIM2 binds to the FAS receptor, either directly or in a complex.
FAS–FAIM2 association increases during 661 W cell apoptosis. 661 W cells were incubated with 500 ng/ml FAS-ligand for various time points to induce apoptosis and FAS was immunoprecipitated. The immunopreciptates were subjected to SDS-PAGE and immunoblotted with FAIM2 antibody, N=4. Graph: Relative band densities were quantified using ImageJ software. The densitometry normalized to untreated cells. Time dependent increase in FAIM2 and FAS interaction showed peak binding (~2-fold) at 12 h after FAS activation. Mean±S.E.M., N=4
FAS activation leads to the phosphorylation of FAIM2 during photoreceptor apoptosis
We have previously shown that JNK and ERK stress kinases are activated after retinal detachment.16 We next evaluated if FAIM2 is phosphorylated following FAS receptor activation using in vitro photoreceptor model 661 W cells. Treatment of 661 W cells with FAS-ligand leads to caspase activation and cell death.6 Similar to photoreceptors after retinal detachment, FAIM2 levels increase in 661 W cells following FAS activation.16 Immunoprecipitation was performed using antibodies specific for phosphorylated Serine (Ser) and Threonine (Thr). Activation of FAS-mediated apoptosis led to increased FAIM2 phosphorylation on Ser and Thr (Figures 6a and b). These findings were confirmed by immunoprecipitating FAIM2 from cells treated with FAS-ligand and demonstrating phosphorylation using antibodies specific for phosphorylated Ser and Thr (Figures 6a and b). These results show that FAIM2 is regulated by phosphorylation during photoreceptor apoptosis.
Phosphorylation of FAIM2 after FAS receptor activation. (a) 661 W cell lysates treated with FAS-ligand (500 ng/ml) were subjected to immunoprecipitation with Phospho-Serine and FAIM2 antibody respectively. Duplicate samples were incubated with λ-phosphatase (1000 units) for 45 min at 30 °C. (b) Immunoprecipitation with Phospho-Threonine and FAIM2 antibody, respectively; N=3
JNK-mediated phosphorylation promotes FAIM2 binding to FAS during photoreceptor apoptosis
Our previous data demonstrated no significant effect on FAIM2 levels in 661 W cells treated with JNK inhibitor SP600125 after exogenous FAS activation.16 We hypothesized that JNK may be regulating photoreceptor survival by secondary modification of FAIM2. To determine if JNK activity is essential for FAIM2 phosphorylation, we repeated phosphorylation analysis of FAIM2 in the presence of JNK inhibitor SP600125. JNK inhibition prevented FAIM2 phosphorylation on Ser and Thr residues following FAS receptor activation (Figure 7a).
JNK activity mediates phosphorylation of FAIM2 after FAS receptor activation. (a) 661 W cell lysates were treated with FAS-ligand (500 ng/ml) with or without JNK inhibitor 10 μM SP600125 (SP) were subjected to immunoprecipitation with FAIM2 antibody after 24 h. Immunoblotting was performed with phospho-Serine (P-Ser), phospho-Threonine (P-Thr), and FAIM2 antibodies. N=3. (b) JNK-mediated FAIM2 phosphorylation is important for FAS–FAIM2 association. 661 W cells were incubated with 500 ng/ml FAS-ligand with or without SP600125 (SP) for 16 h to induce apoptosis and FAS was immunoprecipitated. The immunopreciptates were immunoblotted with FAIM2 antibody, N=3. P-c-JUN immunoblot shows JNK activation after FAS-ligand treatment as measured by c-Jun phosphorylation, which is prevented by SP600125 treatment
Although retinal detachment led to rapid FAIM2/FAS interaction, there was no detectable change in FAIM2 or FAS levels during this time (Figure 5), indicating the potential role of posttranslational FAIM2 modification and increased affinity for FAS binding. When we blocked FAIM2 phosphorylation with JNK inhibition, we saw a significant decrease in FAIM2/FAS interaction following FAS activation (Figure 7b). These results demonstrate that FAIM2 is regulated by JNK-mediated phosphorylation during photoreceptor apoptosis and phosphorylation is important for FAS binding.
Decreased ubiquitination and proteasome-mediated degradation of FAIM2 during FAS-mediated cell death
Our results demonstrate that FAIM2 levels increase in cells immediately after FAS activation but Faim2 mRNA levels remain stable early during cell death. These observations indicate posttranslational regulation of FAIM2 levels in cells. To determine if FAIM2 degradation in cells is mediated by proteasome pathway, we performed cycloheximide (CHX) chase experiments in 661 W cells. When protein synthesis was inhibited, FAIM2 levels decreased in cells over time and close to 50% of FAIM2 was degraded by 24 h (Figures 8a and b). This loss of FAIM2 protein was inhibited by proteasome-inhibitor MG132, indicating that FAIM2 is targeted to the proteasome for degradation in 661 W cells (Figures 8a and b). Similarly, FAS activation in 661 W cells with FAS-ligand treatment blocked FAIM2 degradation, demonstrating that FAIM2 proteasome-targeting is prevented via FAS signaling (Figures 8a and b). Since FAIM2 is phosphorylated by JNK activation during FAS-induced cell death, we determined the effect of phosphorylation on FAIM2 degradation. When JNK activity was inhibited with SP600125, FAS-ligand treatment failed to prevent FAIM2 degradation in cells, demonstrating the importance of JNK-mediated phosphorylation for proteasome targeting of FAIM2 (Figure 8c).
FAS receptor activation prevents proteasome-mediated FAIM2 degradation. (a) 661 W cells were treated 50 μg/ml of cycloheximide (CHX) with or without proteasome inhibitor MG132 (MG), FAS-ligand (Fas-L), or DMSO vehicle and FAIM2 protein level was measured at indicated times by Western blotting. (b) Levels of FAIM2 decreased with time in cells treated with CHX, which was completely reversed by MG132 and partially reversed by Fas-L treatment, N=3, mean±S.E.M. (c) Inhibition of FAIM2 phosphorylation by JNK-inhibitor SP600125 (SP) led to increased proteasome-mediated FAIM2 degradation. N=3, mean±S.E.M.
Ubiquitination of proteins are necessary for proteasome targeting.18 To determine if FAIM2 is ubiquitylated in the retina, we immunoprecipitated FAIM2 from attached and detached rodent retinas and checked for high MW FAIM2 species. We found that FAIM2 is ubiquitylated in the retina under physiologic conditions (Figure 9a). Following retinal detachment, FAIM2 is deubiquitylated, leading to decreased proteasome targeting and increased FAIM2 in the retina after stress (Figure 9a). Changes in FAIM2 ubiquitination and proteasome degradation after detachment was a specific response modulating posttranslational FAIM2 modification, as we did not find a global decrease in protein ubiquitination (Figure 9b) or increase in the levels of GRK, a well-known proteasome substrate in the retina (Figure 9c).19, 20 Similar to in vivo findings, proteasome inhibition resulted in increased FAIM2 ubiquitination in 661 W cells (Figure 9d) and FAS activation did not affect global ubiquitination of proteins in vitro (Figure 9e). These results demonstrate that FAIM2 levels are regulated by proteasome-mediated degradation and JNK-mediated FAIM2 phosphorylation prevents FAIM2 proteasome targeting and leads to FAIM2 accumulation independent of gene transcription.
FAIM2 ubiquitination is regulated by retinal detachment. (a) FAIM2 was immunoprecipitated from attached and detached retinas and Ubiquitin levels were measured by western blotting. In attached retina under normal conditions, high MW complexes representing ubiquitylated FAIM2 (Ub-FAIM2) are seen. Following retinal detachment, ubiquitin-reactive high MW complexes are not seen in FAIM2 immunoprecipitate. (b) Ubiquitin western blot did not demonstrate increased high MW ubiquitin complexes in whole retina extracts after retinal detachment. (c) Proteasome substrate GRK1 levels did not change after retinal detachment. ACTIN is shown as loading control. (d) Inhibition of proteasome by MG132 led to the accumulation of ubiquitylated FAIM2 in 661 W cells. Cells were treated with FAS-ligand (Fas-L) to increase FAIM2 levels and MG132 was included to block proteasome activity. (e) Activation of FAS did not have a detectable effect on proteasome activity in 661 W cells. Cells were treated with Fas-L up to 24 h and extracts were analyzed with Ubiquitin immunoblotting
FAIM2 interacts with pro-apoptotic tumor suppressor protein p53 and HSP90
To identify pro- and anti-apoptotic proteins interacting with FAIM2 during photoreceptor stress, we immunoprecipitated FAIM2 from FAS-ligand treated 661 W cells and analyzed the identities of the interacting proteins via unbiased proteomics. There were 31 and 71 potential protein interactors identified by two independent proteomics analysis (Supplementary Figures 1 and 2). Both p53 and HSP90 showed preferential interaction with FAIM2 following FAS-receptor activation (Figures 10a and b). To confirm the results of proteomics analysis, we evaluated the interaction between FAIM2 and p53 via immunoprecipitation. Activation of FAS receptor by FAS-ligand led to increased association of FAIM2 with p53 (Figure 10c). Similarly, immunoprecipitation demonstrated increased binding of total and phosphorylated HSP90 to FAIM2 during photoreceptor apoptosis (Figure 10d). We also evaluated if FAIM2 phosphorylation was important for HSP90 and FAIM2 interaction by dephosphorylating proteins with a phosphatase. FAIM2 binding to HSP90 as determined by immunoprecipitation was decreased after phosphatase treatment, indicating that this interaction after FAS receptor activation is dependent on FAIM2 phosphorylation. When HSP90 activity was inhibited by geldanamycin treatment, FAIM2 levels did not increase after FAS activation, indicating that HSP90 activity is important for FAIM2 accumulation during photoreceptor apoptosis (Figure 10e).
FAIM2 interacts with p53 and HSP90 after FAS activation. (a,b) Immunoprecipitation of protein lysates from 661 W cells treated with FAS-ligand for 24 h with FAIM2 followed by liquid chromatography–tandem mass spectrometry (LC–MS/MS) proteomic analysis of interactors. The peptide abundance of p53 and HSP90 was quantified using normalized spectral abundance factor (NSAF) analysis. Data is representative of two independent experiments. Significant increase of p53 and HSP90 is observed in the FAIM2 complex of proteins immunoprecipitated after FAS receptor activation. (c) 661 W cells were treated with 500 ng/ml of FAS-ligand and incubated for 24 h. Proteins from whole cell lysate were immunoprecipitated with Faim2 antibody and immunoblotted with p53 antibody. Protein lysates were also immunoprecipitated with p53 antibody and immunoblotted with FAIM2 antibody to demonstrate FAIM2/p53 interaction, N=3. (d) Protein from 661 W cells treated with FAS-ligand for 24 h were immunoprecipitated with FAIM2. HSP90 antibody was used for Western blotting, showing an increase in HSP90/FAIM2 interaction after FAS receptor activation. Incubation with λ-phosphatase showed decreased levels of HSP90/FAIM2 binding, N=3. (e) Protein lysates from 661 W cells treated with FAS-ligand for 24 h with vehicle or 10 μM of HSP90 inhibitor geldanamycin (GD) were analyzed for FAIM2 levels. Inhibition of HSP90 activity with GD led to decreased FAIM2 levels in cells following FAS receptor activation, N=3
FAIM2 expression is important for suppressing pro-death gene transcription in photoreceptors
Our data demonstrates that FAIM2 is an important neuroprotective factor in photoreceptors during periods of retina-RPE separation. To better characterize changes regulated by FAIM2 in photoreceptors, we evaluated the expression of pro-apoptotic gene Fas and necroptosis gene Ripk1, both known to be important in photoreceptor death.5, 21, 22, 23 We found that both Fas and Ripk1 mRNA levels increased in wild type mouse retinas following retinal detachment at 24 h (Figures 11a and b). Quantitative RT-PCR analysis showed that Fas and Ripk1 were expressed at higher levels in retinas isolated from mice lacking FAIM2 protein. The levels of Fas and Ripk1 mRNA transcription in Faim2 knockout retinas before any experimental manipulation were similar to the levels seen in wild-type retinas stressed for 24 h by experimental retinal detachment. When Faim2−/− animals underwent retinal detachment, the transcription of Fas or Ripk1 mRNAs showed little change, in contrast to wild-type animals that showed 3–6 fold increase in gene transcription. Our results indicate that Fas and Ripk1 gene expression may be at maximal level in the absence of FAIM2, demonstrating that FAIM2 is crucial for suppressing the expression of genes regulating apoptosis and necroptosis in the retina.
Fas and Ripk1 mRNA levels in the retina with or without experimental retinal detachment at 24H. Quantitative RT-PCR analysis of Fas and Ripk1 in retinas of wild-type (Faim2+/+) and Faim2 knockout (Faim2−/−) mice with β-actin mRNA serving as control. Relative expression was analyzed by comparative threshold cycle (2−ΔΔct) method. Expression values were represented as fold change over control after normalization with actin. Mean±S.E.M., N=2
Discussion
In this study, we show that FAIM2 is an important neuroprotective protein and inhibits FAS-mediated apoptosis of photoreceptors in the retina. Decreased ubiquitination and proteasome-targeting lead to rapid FAIM2 accumulation after retinal-detachment induced photoreceptor stress. JNK-mediated FAIM2 phosphorylation promotes binding to FAS death receptor to prevent cell death. In the absence of FAIM2 expression, photoreceptors express higher levels of pro-death genes Fas and Ripk1 at baseline and undergo cell death more rapidly after FAS receptor activation. Activation of FAS receptor causes rapid increase in FAIM2 levels in photoreceptor cells as an autoregulatory mechanism of apoptotic FAS receptor signaling.
Previous studies have shown that the activation of the FAS death receptor signaling is the primary mechanism of photoreceptor apoptosis after the separation of the neurosensory retina from the underlying RPE.3, 4, 5 Although photoreceptors have the ability to execute cell death via non-apoptotic pathways including necroptosis, existing evidence indicate that the activation of FAS at the photoreceptor cell membrane is upstream of both apoptotic and non-apoptotic cell death pathways. Therefore, inhibition of FAS receptor, either by extracellular antagonism or prevention of the intracellular death domain activation immediately after FAS-ligand binding, would be expected to provide the most effective method of blocking photoreceptor loss in many retinal diseases.5, 6 Our in vitro and in vivo experiments demonstrated that FAIM2 interacts directly with FAS-receptor complex and this interaction is enhanced during photoreceptor stress. We also show that baseline expression of FAIM2 is important for suppressing the level of Fas and Ripk1 expression in the retina. By reducing the expression of Fas, FAIM2 provides additional time for survival pathways to be dominant in stressed photoreceptors.
Our data suggest that the increase in FAIM2 activity immediately after retina-RPE separation is due to increased protein levels, rather that increased transcription. Intracellular processes that depend on gene transcription require additional time and are not as efficient as the activation or disinhibition of molecular machinery. Under baseline conditions, FAIM2 degradation is mediated by the ubiquitin-proteasome pathway in photoreceptors. During stress, including retinal detachment in vivo or FAS receptor activation in vitro, reduced ubiquitination of FAIM2 results in decreased proteasome targeting and increased protein levels. The stress-induced increase in FAIM2 protein levels appear to also have an immediate negative feedback on Fas gene transcription, which would be expected to regulate the number of FAS receptors at the photoreceptor cell membrane and decrease sensitivity to extracellular stimulation by FAS-ligand. FAIM2 contributes to the down-regulation of pro-death genes in the photoreceptors, presumably to give prosurvival pathways a temporal advantage and prevent the commitment of photoreceptors to an irreversible cell death pathway.
Both in vitro and in vivo experiments confirmed the pro-survival function of FAIM2 during retinal-detachment induced photoreceptor apoptosis. However, FAIM2 expression was not required for the normal development of the retina in vivo or the maintenance of photoreceptors under physiologic conditions. Although both Fas and Ripk1 mRNA transcriptions are increased in the absence of FAIM2, the normal development and aging in the retinas of knockout mice indicate that increased expression of these proapoptotic genes is not sufficient to induce cell death in the absence of other stress signals. One potential mechanism may be the availability of FAS-ligand, which is required to activate the membrane FAS receptor complex. Retinal stress is likely needed for FAS-ligand release into the extracellular matrix in the retina and diffusion into the ONL to activate FAS on photoreceptor membranes. Similarly, in the absence of FAS-ligand mediated activation of FAS death receptor complex, intrinsic survival pathways may overcome the effect of increased Ripk1 expression in Faim2 −/− retinas.
Tumor suppressor p53 responds to cell stress, most importantly DNA damage, and activates apoptosis via intrinsic mitochondrial pathway. The exact role of p53 in retinal stress is highly dependent on the model system being studied.24, 25, 26 Although p53 was shown to play an important role in certain models such as oxidative stress, cell death in other models including light-induced photoreceptor apoptosis is independent of p53 activity. p53 is critical for retinal development and specifically regulates rod photoreceptor survival.26 We found that p53 selectively interacts with FAIM2 during FAS-mediated photoreceptor apoptosis in vitro. The exact role of p53 in FAS-mediated photoreceptor apoptosis is unclear but our data suggests that FAIM2 may inhibit photoreceptor apoptosis by increased p53 binding and sequestration. Activation of p53 has been shown to be important for proapoptotic gene transcription including Fas.27, 28 We show that lack of FAIM2 in vivo leads to increased baseline expression of Fas in the retina. Therefore, FAIM2 could be blocking photoreceptor apoptosis by preventing p53-mediated Fas mRNA transcription in the retina.
HSP90 is member of a large family of molecular chaperones known as the heat shock proteins.29 HSP90 inhibitors are used to increase the sensitivity of tumor cells to apoptosis and are being explored as potential cancer therapeutics.29, 30 We found that FAIM2 interacts with HSP90 during FAS-mediated photoreceptor apoptosis in vitro. HSP90 may be an important chaperone in photoreceptors to increase FAIM2 levels and activity during photoreceptor stress, as the inhibition of HSP90 activity prevented FAS-ligand induced FAIM2 increase in vitro. Several studies have shown that HSP90 inhibitors promote p53-mediated apoptosis in tumor cells via mitochondrial apoptosis pathways.29, 30, 31 p53 activation is critical for the transcription of many pro-apoptotic genes including several Bcl-2 family members and Fas.27, 28, 31
The exact relationship among FAIM2, p53 and HSP90 is unclear but structural analysis of HSP90 and p53 has shown that these two proteins interact with each other in cells in a dynamic manner.32, 33, 34 Interaction between HSP90 and p53 blocks PUMA and BAX-meditated apoptosis.28, 31, 35 Inhibition of HSP90 chaperone complex leads to increased expression of p53 and tumor suppression by p53-dependent apoptosis.31 Our results indicate an intricate relationship between FAIM2, p53 and HSP90 during photoreceptor apoptosis and reveal potential mechanism of neuroprotection by FAIM2 in the retina (Figure 12). We propose that photoreceptor stress, including retina-RPE separation, leads to FAS death receptor activation via FAS-ligand. Increased FAS signaling results in JNK-mediated phosphorylation of FAIM2 and decreased ubiquitination. FAIM2 levels quickly increase due to decreased proteasome-mediated degradation. FAIM2 directly interacts with HSP90, increasing the stability of p53/HSP90 complex and inhibiting p53-mediated Fas transcription and activation of caspases. In the absence of FAIM2, expression of pro-death genes including Fas and Ripk1 are high in photoreceptors at baseline, priming them to respond to FAS receptor activation more robustly and activate cell death more rapidly.
In summary, we demonstrate that photoreceptors utilize FAIM2 as an intrinsic neuroprotective factor to delay FAS-mediated apoptosis during retina-RPE separation. FAIM2 levels in photoreceptors is regulated post-transcriptionally immediately after stress. JNK-mediated phosphorylation of FAIM2 promotes FAS binding. FAIM2 interacts with p53 and HSP90 in response to FAS signaling, providing a potential mechanism of neuroprotection by regulating pro-death gene transcription.
Materials and methods
Ethics statement
All experiments were performed in accordance with the Association for Research in Vision and Ophthalmology (ARVO) Statement for the Use of Animals in Ophthalmic and Vision Research. The protocol was approved by the University Committee on Use and Care of Animals of the University of Michigan (Protocol number: PRO00005497).
Experimental model of retinal detachment
Detachments were created in wild-type C57BL/6 mice and Faim2-knock out mice (B6.129P2-Faim2tm1Dgen/J) obtained from Jackson Laboratories (Bar Harbor, ME, USA) as previously described.4 Briefly, rodents were anesthetized with a mix of ketamine (100 mg/ml) and xylazine (20 mg/ml), and pupils were dilated with topical phenylephrine (2.5%) and tropicamide (1%). A 25-gauge needle was used to create a sclerotomy located 1–2 mm posterior to the limbus with care taken to avoid lens damage. A subretinal injector was introduced through the sclerotomy into the vitreous cavity and then through a peripheral retinotomy into the subretinal space. Sodium hyaluronate (10 mg/ml; Abbott Medical Optics, Abbott Park, IL, USA, Healon OVD) was slowly injected to detach the neurosensory retina from the underlying retinal pigment epithelium. In all experiments, approximately one-third to one-half of the neurosensory retina was detached. Detachments were created in the left eye. The right eye served as the control, with all the steps of the procedure performed, except for introduction of the subretinal injector and injection of the sodium hyaluronate.
TUNEL staining and histology
Mice were euthanatized and the eyes were enucleated at 1, 3 and 7 days after creation of the retinal detachment for TUNEL staining. For histology eyes were enucleated after 1 month and 2 months of retinal detachment. To study the long term effect of age, eyes were enucleated at the age of 6 month and 1 year. Whole eyes were fixed overnight at 4 °C in phosphate-buffered saline with 4% paraformaldehyde (pH 7.4). The specimens were embedded in paraffin and were then placed in a tissue processor (Tissue-Tek II; Sakura, Tokyo, Japan) for standard paraffin embedding. Eyes were then sectioned at a width of 6 μm on a standard paraffin microtome. TUNEL staining was performed on the sections using DeadEnd Colorimetric TUNEL System (Promega Corporation Madison, WI, USA) according to the manufacturer's instructions. TUNEL-positive cells in the outer nuclear layer were counted in a masked fashion. For ONL cell count and retinal area measurements, paraffin sections were stained with 0.5% toluidine blue in 0.1% borate buffer.
Cell counts and retinal area measurements
Retinal images were obtained using Leica DM6000 microscope (Leica Microsystems, Wetzlar, Germany). Photoreceptor cell apoptosis was quantified using ImageJ software as the percentage of total cells in the ONL and the TUNEL positive cells. For toluidine blue–stained specimens, the total number of cells in the ONL were measured using a macro program in ImageJ software. The total area of the ONL and retina (from the outer edge of the ONL to the inner limiting membrane) was measured using ImageJ in high-power field (40 ×) images. Photoreceptor inner and outer segments were not included in the total retinal area measurement, given the variable retraction of these elements after detachment of the neurosensory retina, which does not necessarily correlate with viability of the photoreceptors after reattachment. Normalization of ONL cell count to the total retinal area of each section (i.e., ONL cell count divided by total retinal area) was performed to account for possible differences in angles of sectioning and to allow for inter-sample comparison. Data are represented as mean±S.E.M.
Immunohistochemistry
Eyes from wild type C57BL/6 and Faim2−/− mice were enucleated after 1 and 3 days of retinal detachment and placed in 4% paraformaldehyde overnight at 4 °C. Whole eyes were then placed in a tissue processor (Tissue-Tek II; Sakura, Tokyo, Japan) for standard paraffin embedding. Eyes were then sectioned at a width of 6 μm on a standard paraffin microtome. Immunohistochemistry was performed on sections obtained from paraffin-embedded retinas using standard protocol. Epitope unmasking was accomplished by Proteinase K Antigen Retrieval. The primary antibody for FAIM2 was Rabbit Polyclonal Anti-FAIM2 Antibody (Origene, Rockville, MD, USA, Cat. # TA317786) at a concentration of 5 μg/ml in 1% normal goat serum, 1% BSA in PBST. Secondary antibody concentration was 1:1000 in 3% BSA in phosphate-buffered saline.
Cell culture
The 661 W photoreceptor cell line was generously provided by Dr. Muayyad al-Ubaidi (Department of Cell Biology, University of Oklahoma Health Sciences Center, Oklahoma City, OK, USA).17 The 661 W cell line was maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum, 300 mg/l glutamine, 32 mg/l putrescine, 40 μl/l of β-mercaptoethanol, and 40 μg/l of both hydrocortisone 21-hemisuccinate and progesterone. The media also contained penicillin (90 units/ml) and streptomycin (0.09 mg/ml). Cells were grown at 37 °C in a humidified atmosphere of 5% CO2 and 95% air.
Western blot analysis
Retinas from experimental eyes with detachments and control eyes without detachments were dissected from the RPE-choroid at varying time points, homogenized, and lysed in RIPA Lysis and Extraction Buffer (Catalog number: 89900, Life Technologies Corporation, Grand Island, NY, USA). 1 tablet of protease inhibitor (Complete Mini; Roche Diagnostics, Indianapolis, IN, USA) and 1 tablet of phosphatase inhibitor (PhosSTOP; Roche Diagnostics) per 10 ml were added to the lysis buffer before use to prevent proteolysis and maintain protein phosphorylation. The cellular debris was removed by low speed centrifugation and protein concentrations of supernatants were determined by Pierce BCA Protein Assay Kit (Life Technologies Corporation). Protein samples were separated by SDS NuPAGE Novex 10% gels (Invitrogen, Carlsbad, CA, USA; transferred onto Polyvinylidene difluoride membranes that were blocked in blocking buffer (5% nonfat dry milk in phosphate-buffered saline and 0.1% Tween 20) for 1 h, incubated with primary antibody, washed, and incubated with horse radish peroxidase-conjugated secondary antibody, developed with SuperSignal West Dura Extended Duration Substrate, and image captured digitally on Azure c500 (Azure biosystems, Dublin, CA, USA). Densitometry measurements were performed using Image J software. 661 W cells were passaged, and then plated at 30 000 cells per well in six-well culture dishes in growth media. The cells were treated with 500 ng/ml FasL (Recombinant Mouse Fas Ligand/TNFSF6 Protein Cat#: 6128-SA-025R&D Systems Inc., Minneapolis, MN, USA) and 250 ng/ml HA (Hemagglutinin/HA Peptide Antibody Cat. #: MAB060 R&D Systems Inc., Minneapolis, MN, USA) for 2, 4, 8, 12, 16 and 24 h time points. Cells were harvested at the respective time point and prepared as described for Western analysis. Densitometry measurements were performed using Image J software.36
Phosphorylation and immunoprecipitation assays
661 W cells treated with 500 ng/ml FasL and 250 ng/ml HA was incubated for 8 h, harvested and then lysed using Pierce IP Lysis Buffer, (cat. #: 87787 Life Technologies Corporation) containing only protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN, USA). For lambda phosphatase treatment (Lambda Phosphatase, 100 000 units, sc-200312, Santa Cruz Biotechnology, Inc, Dallas, Texas), 500 μg of protein was added to the 1X lambda phosphatase buffer supplemented with 2 mM MnCl2 and incubated at 30 °C for 45 min. Phosphatase treatments were performed using 2000 units of lambda phosphatase. To demonstrate the role of FAIM2 phosphorylation in protein-protein interactions, we dephosphorylated FAIM2 before performing immunoprecipitation. After dephosphorylation, lambda phosphatase treated and control lysate samples were incubated at 4 °C overnight, with 50 μl of Dynabeads protein G suspension (Dynabeads Protein G Immunoprecipitation Kit, 10007D, Life Technologies Corporation) that was preincubated with FAIM2, Phospho-Serine and Phospho-Threonine antibody for 1 h at room temperature. After incubation, the supernatant was collected after separation from the beads with a magnetic device. The bead-antibody-antigen complex was washed to remove any non-specific binding and the antibody-antigen complex was eluted with 30 μl of premixed Nupage LDS sample buffer and elution buffer. The following antibodies were used for immunoprecipitation: Anti-FAIM2 Antibody (Cat. #TA317786 OriGene Technologies, Rockville, MD, USA), Phospho-Serine, (Cat. 344911M, Invitrogen), and Phospho-Threonine (Cat. #9386, Cell Signaling Technology, Danvers, MA, USA).
CHX chase assays
CHX (50 μg/ml) was added to 661 W cells to inhibit protein synthesis and FAIM2 western blot was performed from cell lysates to measure protein degradation at 4, 8 and 24 h with or without 10 μM of proteasome inhibitor MG132. 2 mM of N‐ethylmaleimide (NEM) was included in the cell lysis buffer to inhibit isopeptidases. In some experiments, cells were pretreated with FAS-ligand to induce FAIM2 expression and CHX chase assays were performed in the presence of JNK inhibitor SP600125 (10 μM).
Quantitative real-time PCR
Total RNA was purified from attached and detached retinal tissues of C57/BL6 (Faim2 +/+) and Faim2-knock out (Faim2−/−) mice using RNeasy Mini kit (Qiagen, Germantown, MD, USA, Cat No./ID: 74104) and QIAshredder (Qiagen, Cat No./ID: 79654). cDNA was synthesized using RT2 First Strand Kit (Cat. No./ID: 330401) by reverse transcription using random hexamers and oligo-dT. Triplicate qPCRs were performed using TaqMan™ Universal PCR master mix (Applied Biosystems, Thermo fisher Scientific). Reactions were performed and monitored using a CFX96 real time PCR system (Bio-Rad Laboratories), 1 cycle at 50 °C for 2 min, 1 cycle at 95 °C for 10 min followed by 39 cycles at 95 °C for 15 s, 60 °C for 1 min. Relative normalized mRNA fold changes were calculated using the 2^(−ddct) method.
Immunoprecipitation profile of FAIM2 by proteomics
661 W cells treated with FasL and HA for 8 h were lysed with Pierce immunoprecipitation assay (IPA) buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40 and 5% glycerol) containing protease inhibitor cocktail and Phosphostop. The total protein concentration of the cell lysate was determined using a Micro BCA protein assay kit (Thermoscientific, Rockford, IL, USA). 1 mg cell lysate and Dynabeads® Protein G (Cat #: 10004D, Thermo fisher) were used for immunoprecipitation. 1 mg cell lystate was suspended in the precleared Dynabeads–FAIM2 antibody complex and incubated overnight at 4 °C with rocking. The immunoprecipitated protein (Dynabead-Ab-Ag complex) was then analyzed with unbiased proteomics to identify interacting partners. Briefly, the procedure involves SDS-PAGE, band excision, in-gel digestion with trypsin, LC/MS/MS. Normalized spectral abundance factor (NSAF) was calculated to identify the proteins interacting with FAIM2 preferentially during Fas-induced apoptosis. Raw data from two independent experiments are included in Supplementary Figures 1 and 2.
Antibodies
LFG (Cat. #: AS-54488, Anaspec, Fremont, CA, USA), Phosphothreonine (Cat. #: AB1607, EMD Millipore, Billerica, MA, USA), Phosphoserine (Cat. #: AB1603, EMD Millipore), p53 (Cat. #: AF1355, R&D Systems, Minneapolis, MN, USA), Phospho-HSP90α (Thr5/7) (Cat. #3488, Cell Signaling Technology), HSP90 (Cat.#4874, Cell Signaling Technology), β-Actin (A5316-100UL, Sigma-Aldrich, St. Louis, MO, USA), Ubiquitin PD-40 (Cat.#, Cell Signaling Technology). Secondary antibodies were anti- rabbit, anti-mouse, or anti-goat IgG-HRP.
Data analysis
Results are expressed as mean±S.E.M. Data was analyzed using Student t-test or one-way ANOVA followed by Bonferroni post hoc test. A value of P<0.05 was considered significant. Prism 6.0 (GraphPad Software, San Diego, CA, USA) was used for all statistical analysis.
References
Piccolino FC, de la Longrais RR, Ravera G, Eandi CM, Ventre L, Abdollahi A et al. The foveal photoreceptor layer and visual acuity loss in central serous chorioretinopathy. Am J Ophthalmol 2005; 139: 87–99.
Burton TC . Recovery of visual acuity after retinal detachment involving the macula. Trans Am Ophthalmol Soc 1982; 80: 475–497.
Zacks DN, Hanninen V, Pantcheva M, Ezra E, Grosskreutz C, Miller JW . Caspase activation in an experimental model of retinal detachment. Invest Ophthalmol Vis Sci 2003; 44: 1262–1267.
Zacks DN, Zheng QD, Han Y, Bakhru R, Miller JW . FAS-mediated apoptosis and its relation to intrinsic pathway activation in an experimental model of retinal detachment. Invest Ophthalmol Vis Sci. 2004; 45: 4563–4569.
Zacks DN, Boehlke C, Richards AL, Zheng QD . Role of the Fas-signaling pathway in photoreceptor neuroprotection. Arch Ophthalmol. 2007; 125: 1389–1395.
Besirli CG, Chinskey ND, Zheng QD, Zacks DN . Inhibition of retinal detachment-induced apoptosis in photoreceptors by a small peptide inhibitor of the fas receptor. Invest Ophthalmol Vis Sci 2010; 51: 2177–2184.
Ross WH . Visual recovery after macula-off retinal detachment. Eye 2002; 16: 440–446.
Ross WH, Stockl FA . Visual recovery after retinal detachment. Curr Opin Ophthalmol 2000; 11: 191–194.
Chong DY, Boehlke CS, Zheng QD, Zhang L, Han Y, Zacks DN . Interleukin-6 as a photoreceptor neuroprotectant in an experimental model of retinal detachment. Invest Ophthalmol Vis Sci 2008; 49: 3193–3200.
Zacks DN . Gene transcription profile of the detached retina (An AOS Thesis). Trans Am Ophthalmol Soc 2009; 107: 343–382.
Hu L, Smith TF, Goldberger G . LFG: a candidate apoptosis regulatory gene family. Apoptosis 2009; 14: 1255–1265.
Somia NV, Schmitt MJ, Vetter DE, Van Antwerp D, Heinemann SF, Verma IM . LFG: an anti-apoptotic gene that provides protection from Fas-mediated cell death. Proc Natl Acad Sci USA 1999; 96: 12667–12672.
Beier CP, Wischhusen J, Gleichmann M, Gerhardt E, Pekanovic A, Krueger A et al. FasL (CD95L/APO-1L) resistance of neurons mediated by phosphatidylinositol 3-kinase-Akt/protein kinase B-dependent expression of lifeguard/neuronal membrane protein 35. J Neurosci 2005; 25: 6765–6774.
Reich A, Spering C, Gertz K, Harms C, Gerhardt E, Kronenberg G et al. Fas/CD95 regulatory protein Faim2 is neuroprotective after transient brain ischemia. J Neurosci 2011; 31: 225–233.
Hurtado de Mendoza T, Perez-Garcia CG, Kroll TT, Hoong NH, O'Leary DD, Verma IM . Antiapoptotic protein Lifeguard is required for survival and maintenance of Purkinje and granular cells. Proc Natl Acad Sci USA 2011; 108: 17189–17194.
Besirli CG, Zheng QD, Reed DM, Zacks DN . ERK-mediated activation of Fas apoptotic inhibitory molecule 2 (Faim2) prevents apoptosis of 661W cells in a model of detachment-induced photoreceptor cell death. PloS One 2012; 7: e46664.
al-Ubaidi MR, Font RL, Quiambao AB, Keener MJ, Liou GI, Overbeek PA et al. Bilateral retinal and brain tumors in transgenic mice expressing simian virus 40 large T antigen under control of the human interphotoreceptor retinoid-binding protein promoter. J Cell Biol 1992; 119: 1681–1687.
Campello L, Esteve-Rudd J, Cuenca N, Martin-Nieto J . The ubiquitin-proteasome system in retinal health and disease. Mol Neurobiol 2013; 47: 790–810.
Aguila M, Bevilacqua D, McCulley C, Schwarz N, Athanasiou D, Kanuga N et al. Hsp90 inhibition protects against inherited retinal degeneration. Hum Mol Genet 2014; 23: 2164–2175.
Penela P . Chapter three - ubiquitination and protein turnover of G-protein-coupled receptor kinases in GPCR signaling and cellular regulation. Prog Mol Biol Transl Sci 2016; 141: 85–140.
Trichonas G, Murakami Y, Thanos A, Morizane Y, Kayama M, Debouck CM et al. Receptor interacting protein kinases mediate retinal detachment-induced photoreceptor necrosis and compensate for inhibition of apoptosis. Proc Natl Acad Sci USA 2010; 107: 21695–21700.
Zadro-Lamoureux LA, Zacks DN, Baker AN, Zheng QD, Hauswirth WW, Tsilfidis C . XIAP effects on retinal detachment-induced photoreceptor apoptosis [corrected]. Invest Ophthalmol Vis Sci 2009; 50: 1448–1453.
Murakami Y, Miller JW, Vavvas DG . RIP kinase-mediated necrosis as an alternative mechanisms of photoreceptor death. Oncotarget 2011; 2: 497–509.
Vuong L, Brobst DE, Saadi A, Ivanovic I, Al-Ubaidi MR . Pattern of expression of p53, its family members, and regulators during early ocular development and in the post-mitotic retina. Invest Ophthalmol Vis Sci 2012; 53: 4821–4831.
Vuong L, Conley SM, Al-Ubaidi MR . Expression and role of p53 in the retina. Invest Ophthalmol Vis Sci 2012; 53: 1362–1371.
Vuong L, Brobst DE, Ivanovic I, Sherry DM, Al-Ubaidi MR . p53 selectively regulates developmental apoptosis of rod photoreceptors. PloS One 2013; 8: e67381.
Schilling T, Schleithoff ES, Kairat A, Melino G, Stremmel W, Oren M et al. Active transcription of the human FAS/CD95/TNFRSF6 gene involves the p53 family. Biochem Biophys Res Commun 2009; 387: 399–404.
Wilson AM, Morquette B, Abdouh M, Unsain N, Barker PA, Feinstein E et al. ASPP1/2 regulate p53-dependent death of retinal ganglion cells through PUMA and Fas/CD95 activation in vivo. J Neurosci 2013; 33: 2205–2216.
Solarova Z, Mojzis J, Solar P . Hsp90 inhibitor as a sensitizer of cancer cells to different therapies (review). Int J Oncol 2015; 46: 907–926.
Regan PL, Jacobs J, Wang G, Torres J, Edo R, Friedmann J et al. Hsp90 inhibition increases p53 expression and destabilizes MYCN and MYC in neuroblastoma. Int J Oncol 2011; 38: 105–112.
He K, Zheng X, Zhang L, Yu J . Hsp90 inhibitors promote p53-dependent apoptosis through PUMA and Bax. Mol Cancer Ther 2013; 12: 2559–2568.
Hagn F, Lagleder S, Retzlaff M, Rohrberg J, Demmer O, Richter K et al. Structural analysis of the interaction between Hsp90 and the tumor suppressor protein p53. Nat Struct Mol Biol 2011; 18: 1086–1093.
Park SJ, Borin BN, Martinez-Yamout MA, Dyson HJ . The client protein p53 adopts a molten globule-like state in the presence of Hsp90. Nat Struct Mol Biol 2011; 18: 537–541.
Park SJ, Kostic M, Dyson HJ . Dynamic interaction of Hsp90 with its client protein p53. J Mol Biol 2011; 411: 158–173.
Matsumoto H, Murakami Y, Kataoka K, Lin H, Connor KM, Miller JW et al. Mammalian STE20-like kinase 2, not kinase 1, mediates photoreceptor cell death during retinal detachment. Cell Death Dis 2014; 5: e1269.
Abramoff MD, Magalhaes PJ, Ram SJ . Image processing with ImageJ. Biophotonics Int 2004; 11: 36–42.
Acknowledgements
Supported by grants from the NEI 5K08EY023982-03, Research to Prevent Blindness, and the Departmental Core Center for Vision Research EY-07003. We thank Tam-Thien Hoang for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Dr Besirli and Dr Zacks have intellectual property interests and are listed as inventors on a patent for the use of FAIM2 as a retinal neuroprotective treatment. The remaining authors declare no conflict of interest.
Additional information
Edited by S Nagata
Supplementary Information accompanies this paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Pawar, M., Busov, B., Chandrasekhar, A. et al. FAS apoptotic inhibitory molecule 2 is a stress-induced intrinsic neuroprotective factor in the retina. Cell Death Differ 24, 1799–1810 (2017). https://doi.org/10.1038/cdd.2017.109
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2017.109
- Springer Nature Limited
This article is cited by
-
Small molecule activation of metabolic enzyme pyruvate kinase muscle isozyme 2, PKM2, circumvents photoreceptor apoptosis
Scientific Reports (2020)
-
Hexokinase 2 is dispensable for photoreceptor development but is required for survival during aging and outer retinal stress
Cell Death & Disease (2020)
-
Cell death pathologies: targeting death pathways and the immune system for cancer therapy
Genes & Immunity (2019)