Abstract
Background:
MicroRNA expression signatures can promote personalised care for non-small cell lung cancer (NSCLC) patients. Our aim was to evaluate the previously unexplored prognostic potential of miR-197, a key oncogenic molecule for NSCLC.
Methods:
Total RNA isolation (n=124 NSCLC and n=21 tumour-adjacent normal tissues), was performed using the QIAsymphony SP workstation. The quantity and quality of RNA were assessed by spectrophotometric analysis and an Agilent 2100 bioanalyzer. Polyadenylation and reverse transcription were subsequently carried out. MiR-197 expression levels were measured by qPCR, after quality control (inter-assay CV=7.8%). Internal validation procedures were followed by assigning training and test sets and robust biostatistical analyses were performed, including bootstrap resampling.
Results:
MiR-197 is associated with larger tumours (P=0.042) and the squamous cell carcinoma histotype (P=0.032). Interestingly, after adjusting for important prognostic indicators, miR-197 expression was identified as a novel independent predictor of unfavourable prognosis for NSCLC patients (HR=1.97, 95% CI=1.10–3.38, P=0.013). We also demonstrate that miR-197 retains its prognostic performance in both early-stage I (P=0.045) and more advanced-stage individuals (P=0.036).
Conclusions:
The cost-effective expression analysis of miR-197 could constitute a novel molecular tool for NSCLC management.
Similar content being viewed by others
Main
Optimal confrontation of non-small cell lung cancer (NSCLC) remains a largely unachieved objective, as the 5-year survival rate struggles to surpass 15% (Powell et al, 2013; Reck et al, 2013). Most decisions throughout the clinical management of NSCLC are dictated by TNM staging. Surgical removal of tumours, where feasible, is the gold standard of treatment (Peters et al, 2012; Reck et al, 2013; Vansteenkiste et al, 2013; Johnson et al, 2014). However, even early-stage resected patients present an extremely variable oncologic outcome; this is a reflection of the large biological heterogeneity between patients who are currently erroneously categorised in the same prognostic group (Powell et al, 2013). For example, adjuvant chemotherapy has not been proven to prolong the survival of stage I patients (especially stage IA; Gadgeel et al, 2012; Reck et al, 2013; Johnson et al, 2014). Meanwhile, ∼30–40% of stage I NSCLC patients are expected to relapse within 5 years (Martini et al, 1995; Goodgame et al, 2008; Zhu et al, 2010), and this illustrates a severe inability, intrinsic to TNM staging, in defining high-risk patients who should receive adjuvant chemotherapy. In advanced disease stages, an accurate prognostic indicator could guide the administration of maintenance therapy, a clinical decision that currently remains under serious controversy (Gadgeel et al, 2012; Peters et al, 2012; Johnson et al, 2014). Consequently, there is a necessity for novel molecular prognostic indicators that would enhance the conventional staging system and improve the decision-making process.
MicroRNAs (miRNAs) are regarded as a novel source of biomarkers for the majority of human malignancies (Nana-Sinkam and Croce, 2013) and NSCLC is not an exception (Vannini et al, 2013). From the very first study, conducted in 2004 by Takamizawa et al, addressing the prognostic value of let-7 in NSCLC (Takamizawa et al, 2004), up to contemporary research, miRNAs are proving to have a decisive role in lung cancer pathobiology and to represent promising molecular markers for NSCLC prognosis (Boeri et al, 2012; Shen and Jiang, 2012; Vannini et al, 2013; Liloglou et al, 2014).
MicroRNA 197 (miR-197) represents a molecule that is associated with a wide range of pathologic conditions ranging from non-neoplastic diseases (e.g., type 2 diabetes, myocardial infarction, metabolic syndrome) to major human malignancies (e.g., NSCLC, hepatocellular carcinoma, pancreatic cancer, thyroid cancer; detailed overview in Table 1). Interestingly, a recent study by Fiori et al (2014) provided strong evidence that miR-197 is a functional oncomiR with a central role in NSCLC pathogenesis, mediated through the suppression of the p53-dependent apoptotic cascade. MiR-197 targeting is therapeutically relevant, as it can inhibit tumour growth in vivo (Fiori et al, 2014). Previous studies had also shown that miR-197 regulates the expression of the tumour suppressor FUS1 in NSCLC and is upregulated in the tissue and plasma of lung cancer patients (Du et al, 2009; Zheng et al, 2011).
Consequently, based on: (i) the robust association between miR-197 and NSCLC pathogenesis/progression (Du et al, 2009; Fiori et al, 2014, ii) the key oncogenic role of miR-197 in aggressive human malignancies (Hamada et al, 2013; Dai et al, 2014) and (iii) the diagnostic capacity of circulating miR-197 in lung cancer (Zheng et al, 2011; Abd-El-Fattah et al, 2013), we aimed to evaluate the previously unexplored potential of this lung oncomiR as a novel prognostic tissue biomarker for NSCLC.
Materials and methods
Collection, storage and characteristics of the NSCLC tissue specimens
A total of 124 primary NSCLC tissue specimens, as well as 21 samples from normal tumour-adjacent tissue parts, were collected from surgically resected patients with early and advanced disease stages at the Trousseau Hospital, Tours, France, between 2006 and 2011. The mean age±s.e. of patients was 65.3±0.94. No neoadjuvant treatment was administered in these patients. All the participating patients gave their informed consent before the initiation of this study, which was designed and performed following the ethical guidelines of the Helsinki Declaration and the French bioethical committees.
Tissue samples were selected and reviewed by an experienced pathologist. The non-cancerous parts had a distance of at least 3 cm from the tumour. Upon selection, the samples were quickly frozen under liquid nitrogen and stored for further analysis at −80 °C. Histological diagnosis and tumour grading was established according to the World Health Organisation classification of lung tumours. Tumour staging was performed in agreement with the seventh lung cancer TNM classification and staging system. Detailed clinical and pathological characteristics of the patients collective are described in Table 2.
Tissue homogenisation, total RNA isolation, polyadenylation and reverse transcription
The Tissue-Tek O.C.T. (Sakura Finetek Europe, Alphen aan den Rijn, The Netherlands) and a cryotome (Thermo Scientific Inc., Waltham, MA, USA) platform were used for sectioning the fresh frozen NSCLC tissue samples (⩽50 mg tissue per sample). Consequently, the tissue parts were lysed in 430 μl lysis buffer (RLT Plus, Qiagen Inc., Valencia, CA, USA) with 143 μM β-mercaptoethanol and, upon complete homogenisation, were centrifuged for 1 min at maximum speed. Total RNA was isolated from the resulting supernatant using QIAsymphony SP workstation and the RNA CT 400 protocol (Qiagen Inc.). To avoid DNA contamination, digestion with DNAse I was also performed. The quantity, quality and purity of the resulting RNA were thoroughly assessed by a NanoDrop 2000c spectrophotometer (Thermo Scientific Inc.) and an Agilent 2100 bioanalyzer. Only intact RNA samples with RIN>6 were considered for downstream applications.
Polyadenylation and reverse transcription of total RNA, including miRNAs, was performed as previously described (Mavridis et al, 2013). In brief, 500 ng of RNA was polyadenylated in the presence of ATP (800 μ M) by 1 U of E. coli Poly(A) Polymerase in the reaction buffer supplied by the manufacturer (New England Biolabs Inc., Ipswich, MA, USA) at 37 °C for 60 min, followed by an enzyme inactivation step at 65 °C for 10 min. Immediately after that, the polyadenylated RNA was reverse transcribed into first-strand cDNA in a 20 μl reaction containing the reaction buffer of the manufacturer, 100 U M-MLV reverse transcriptase in the reaction, 20 U RNaseOUT recombinant ribonuclease inhibitor (Invitrogen, Carlsbad, CA, USA) and 0.25 μ M poly(T) adapter (5′-GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN-3′) at 37 °C for 60 min, followed by an enzyme inactivation step at 70 °C for 15 min.
Quantitative PCR for miR-197 expression analysis
MiR-197-3p expression levels were quantified using a SYBR-Green-based qPCR assay in a 10-μl reaction, which was run in duplicates in 96-well fast reaction plates (Applied Biosystems, Carlsbad, CA, USA). The reactions consisted of Kapa SYBR Fast Universal qPCR Master Mix (Kapa Biosystems, Wilmington, MA, USA) including Rox Low passive reference dye, a specific miR-197-3p- (5′-CACCACCTTCTCCACCCA-3′) or a SNORD48- (5′-TGATGATGACCCCAGGTAACTCT-3′) specific forward primer and a universal reverse primer (5′-GCGAGCACAGAATTAATACGAC-3′), all at a final concentration of 200 nM, as well as 1 ng of cDNA as template. The sizes of the miR-197 and SNORD48 amplicons were 62 and 105 bp, respectively. The qPCR reactions took place in a 7500 Fast Real-Time PCR System (Applied Biosystems) using the following rapid cycling thermal protocol: a 3-min polymerase activation step at 95 °C and 40 cycles of denaturation — annealing/extension steps at 95 °C for 3 s — 60 °C for 30 s. The melting curve analysis protocol followed immediately after. Each run included a no-template control. MiR-197 expression levels were normalised to the endogenous reference control SNORD48 using the formula ΔCT=CTmiR-197−CTSNORD48. To allow inter-run comparisons, the same designated cancerous sample (calibrator) was included in all qPCR runs. MiR-197 relative quantification (RQ) expression units were calculated relatively to the calibrator via the formula RQ=2−ΔΔCT, where ΔΔCT=ΔCTsample−ΔCTcalibrator, by the 7500 software v.2.06 (Applied Biosystems).
Quality control of the developed qPCR method
The developed qPCR method went through quality control to verify its specificity, sensitivity and reproducibility. To test the specificity of the reactions, melting curve analysis and agarose gel electrophoresis were performed (Supplementary Figure S1). The presence of a unique peak in the melting curve and a unique band detected after 3.0% agarose gel electrophoreses corroborated that a single specific amplicon was produced for each case throughout the qPCR reactions.
We also assayed a series of negative control samples, including no-template controls, enzyme-negative controls (reverse transcriptase, poly(A)polymerase), as well as DNA template controls, all of which failed to generate a detectable CT value.
Standard curves were performed for both the endogenous and target molecules, by using a series of cDNA dilutions covering 5 orders of magnitude (r2=0.999 for SNORD48 and r2=0.998 for miR-197). PCR efficiencies were calculated using the slopes of the standard curves equal to 95% for SNORD48 and 100% for miR-197 (Supplementary Figure S1); the presence of similar qPCR efficiencies that are both under the acceptable range allows the use of the comparative CT method and excludes any PCR inhibition.
The method’s reproducibility was assessed by analysing common samples (n=7) in different qPCR runs and calculating the coefficient of variation from duplicate measurements (Jones and Payne, 1997). The inter-assay CV was found to be 7.8%.
Biostatistical analyses – internal validation
Spearman’s correlation analysis was used to analyse the associations between the continuous variables of the study. The analysis of differences in miR-197 expression levels between distinct groups of NSCLC patients was performed via the Mann–Whitney U-test or the Jonckheere–Terpstra test, where applicable. Paired samples analysis was conducted according to Wilcoxon signed-rank test.
The X-tile algorithm (Camp et al, 2004) was used to classify patients into miR-197-high- and miR-197-low-expression groups. More precisely, X-tile was utilised to select an optimal cutoff point while correcting for the use of minimum P-value statistics. Two robust methods of P-value correction were used that produced a cutoff with minimal chance of type I errors: (i) the Miller–Siegmund P-value correction and (ii) the Monte-Carlo simulations by cross-validation and the corrected P-value approaches based on 1000 random populations. Internal validation was also performed by using the X-tile option for automatically generating random training and validation cohorts, finding the optimal cutoff point in the training set and applying this cutoff to the validation cohort; a similar procedure was also followed by creating random training and validation sets via the SPSS software. All the aforementioned methods produced the same optimal cutoff point, that is, 0.635 RQ units of miR-197 expression (61st percentile).
After establishing and internally validating the robustness of the selected cutoff, survival analysis was performed by generating the Kaplan–Meier overall survival (OS) curves to evaluate the prognostic potential of miR-197 expression for NSCLC patients. Cox regression models were developed at the univariate and multivariate levels. Multivariate Cox regression was performed with the purpose of testing for independence of the miR-197-driven prognostic information, adjusting for potentially confounding variables via a standardised and a final model. The standardised model included the marker under evaluation (miR-197) along with standard prognostic variables (tumour size, nodal status, pTNM stage, differentiation, histotype), as well as typical patient demographic variables (age and gender); this standardised model can facilitate the comparison of the prognostic performance of miR-197 across the present and future studies. The final multivariate model included the variables that were found to be robustly associated with OS in univariate analysis at a statistically significant level (i.e., miR-197, pTNM stage and differentiation status). The robustness of all the above-mentioned models was validated by the bootstrap approach. Random sampling was performed with replacement, producing 1000 bootstrap samples; the bias corrected and accelerated approach was followed for the calculation of 95% confidence interval (CI).
We then followed a complementary statistical approach, based on the stratification of patients into subgroups, and the application of the Kaplan–Meier survival analyses for miR-197 expression in these groups. This approach was followed to evaluate the prognostic performance of miR-197 in meaningful subgroups of patients and irrespective of different treatments.
All the aforementioned statistical analyses were performed using the X-tile v3.6.1 (Yale University, New Haven, CT, USA), SPSS Statistics v20.0 (IBM Corp., Armonk, NY, USA) and MedCalc v.12.5 (MedCalc Software, Ostend, Belgium) software. Two-tailed tests were used and P-values <0.05 were adapted for statistical significance.
Results
The expression of miR-197 in NSCLC tissues is associated with larger tumour size, and squamous cell carcinoma tumours
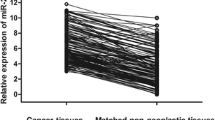
By analysing 21 cancerous and tumour-adjacent normal tissue pairs, no statistically significant difference was observed regarding miR-197 expression (P=0.357). The expression levels of miR-197 in the 124 NSCLC tissue samples analysed ranged from 0.159 to 2.46 RQ units, with a median value of 0.512 and a mean±s.e.=0.618±0.035.
The expression of miR-197 in cancer was positively associated with tumour size, as it was increased in patients harbouring tumours >3 cm compared with patients with ⩽3 cm tumours (P=0.042). MiR-197 levels were also elevated in squamous cell carcinoma compared with adenocarcinoma (P=0.032; Figure 1). A weak, positive correlation was also observed between the expression levels of miR-197 and patients’ age (rs=0.178, P=0.048), whereas no significant associations were observed between miR-197 expression and pTNM stage (P=0.496), tumour differentiation (P=0.141), lymph node status (P=0.951), metastasis (P=0.910), smoking status (P=0.371), pack-years (P=0.826), the presence of chronic obstructive pulmonary disease (P=0.589) or gender (P=0.093).
Distribution of miR-197 expression in NSCLC tissues. Distribution of miR-197 expression in NSCLC tissues of different tumour size (A) and histotype (B). ADC=adenocarcinoma, SCC=squamous cell carcinoma. P-value calculated by the Mann–Whitney U-test. y axis is in logarithmic scale. Bold lines represent the median value.
Stratification to high- and low-risk groups based on miR-197 expression: selection of an optimal cutoff value and internal validation
An appropriate cutoff (equal to 0.635 RQ units) that was able to effectively stratify our cohort, with respect to OS probabilities, was selected while correcting the chance of type I errors (Miller–Siegmund P=0.022). Monte-Carlo simulations in 1000 random populations resulted in the same, statistically significant results (cross–validation P=0.027, Monte-Carlo corrected P=0.031). The aforementioned cutoff value was identified in the X-tile randomly generated training cohort as optimal (P=0.034) and then its validity was confirmed in the validation cohort (P=0.012). The same results were obtained by creating random training and validation cohorts via SPSS (optimal cutoff=0.635, P=0.030 for the training and P=0.004 for the validation cohort, according to the Kaplan–Meier analysis).
MiR-197 is robustly associated with adverse oncologic outcome
Complete follow-up data regarding the OS of NSCLC patients were available for the whole cohort (n=124). Median follow-up time was 27.5 months (1.0–81 months); median follow-up in patients still alive at the time of analysis was 40 months (14–81 months).
As indicated by the Kaplan–Meier survival analysis, patients categorised as miR-197-high clearly presented (P=0.0007) a worse OS course compared with the miR-197-low ones (Figure 2A). The median survival time for miR-197-low individuals was 61 months, which is in striking difference with the corresponding period of 23 months for miR-197-high patients. Furthermore, when focusing on the 1-year survival time point, it was demonstrated that miR-197 levels were considerably higher among the deceased patients (median=0.740 RQ units) by 1 year compared with the survivors (median=0.490 RQ units) at 1 year (P=0.017; Figure 2B).
Association of miR-197 expression with unfavourable prognosis in the whole cohort of NSCLC patients. (A) Kaplan–Meier OS curves for the whole cohort of NSCLC patients, classified as miR-197-high and miR-197-low. P-value calculated by the log-rank test. (B) MiR-197 expression levels in NSCLC patients deceased by 1 year and alive at 1 year post surgery. P-value calculated by the Mann–Whitney U-test. Bold lines represent the median value. *Outliers.
The above-mentioned prognostic potential of miR-197 was corroborated (P=0.001) by univariate Cox proportional hazard regression analysis with bootstrap resampling. Patients stratified as miR-197-high were 2.31 times more likely (95% CI=1.44–3.73) to die compared with miR-197-low ones. As expected, advanced pTNM stage (HR=1.89, 95% CI=1.21–3.23, P=0.011), and poor tumour differentiation (HR=2.12, 95% CI=1.24–3.84, P=0.006), also emerged as significant indicators of adverse oncologic outcome (Table 3).
MiR-197: a novel independent predictor of unfavourable prognosis for NSCLC patients
By using multivariate Cox regression analysis, adjusted for standard prognostic indicators and typical demographic factors, it was demonstrated that miR-197 expression is a strong and independent predictor (P=0.007) of adverse oncologic outcome (HR=2.52, 95% CI=1.23–6.21) for NSCLC patients (Table 3). Other important predictors were pTNM stage (HR=2.83, 95% CI=1.08–8.69, P=0.014) and differentiation status (HR=2.20, 95% CI=1.09–5.52, P=0.023; Table 3).
All statistically significant predictors (miR-197, tumour stage and differentiation) were included in a final model, in which only miR-197 retained its robust prognostic performance (HR=1.97, 95% CI=1.10–3.38, P=0.013; Table 3).
Tissue miR-197 expression is associated with unfavourable prognosis in subgroups of NSCLC patients
As demonstrated in Figure 3, miR-197 expression is significantly associated with adverse oncologic outcome in both stage I (P=0.045) and more advanced stages (II–IV; P=0.036; Figure 3A and B, respectively). These results were confirmed by univariate Cox regression, which showed that stage I patients with high miR-197 expression presented a poorer OS course compared with patients categorised as miR-197-low (HR=2.34, 95% CI=0.974–5.64, P=0.044). For stage II–IV patients, the corresponding HR value was 1.93 (95% CI=1.06–3.90, P=0.038).
In addition, miR-197 showed a statistically significant ability for risk stratification of NSCLC patients regarding OS, irrespectively of the administration of post-operative chemotherapy (P=0.042), Figure 3C and HR=1.95, 95% CI=0.99–4.15, P=0.048) or not (P=0.045, Figure 3D and HR=2.23, 95% CI=0.935–5.47, P=0.045).
Discussion
Contemporary research has proven that molecular signatures can constitute the first, yet decisive, step towards the realisation of personalised cancer care. Successful examples include the routinely used gene expression tests MammaPrint and OncotypeDx for breast cancer (Duffy and Crown, 2008) and the recently FDA-approved multifactorial biomarker panels ROMA score and OVA1 for ovarian cancer (Leung et al, 2012). Novel molecular prognostic indicators, such as miRNAs, could also aid towards enhancing the clinical management of NSCLC patients (Lin and Beer, 2012; Powell et al, 2013). The advantage of miRNAs as prognostic biomarkers include: (i) their reliable and cost-efficient quantification in specimens accompanied by invaluable clinical information such as FFPE tissues, as well as in minimal diagnostic tissue specimens from biopsies (Boeri et al, 2012; Shen and Jiang, 2012; Liloglou et al, 2014, ii) the enhanced clinical information regarding molecular cancer pathogenesis that can derive from their expression analysis; an alteration in the expression of a single miRNA molecule usually reflects changes affecting a broad range of biological pathways (Boeri et al, 2012; Nana-Sinkam and Croce, 2013). MiRNAs are currently investigated as biomarkers for several human malignancies, including NSCLC, in >100 clinical trials (Nana-Sinkam and Croce, 2013).
In the present study we show that miR-197 is robustly associated with an adverse OS outcome in NSCLC patients, independently of several important prognostic indicators such as pTNM stage (Table 3). After analysing meaningful NSCLC patient subgroups, we demonstrate that miR-197 retains its predictive properties in stage I, as well as in more advanced disease stages (Figure 3). Interestingly, miR-197 has already been evaluated as a circulating diagnostic marker for NSCLC; plasma and serum measurements of miR-197 can distinguish lung cancer patients from healthy individuals (Zheng et al, 2011; Abd-El-Fattah et al, 2013) and when they are used in combination with other miRNA molecules are predictive of lung cancer risk and the presence of aggressive disease (Boeri et al, 2011) (Table 1).
The apparent association between tissue miR-197 expression and adverse oncologic outcome, described in the present study, is in fine agreement with its previously reported oncogenic role in lung cancer (Du et al, 2009; Fiori et al, 2014). More precisely, it has been recently proven that miR-197 exerts its lung tumour-promoting role by inhibiting the p53 apoptotic cascade, through a mechanism that includes the suppression of NOXA and BMF genes; interestingly, in vivo experiments showed that miR-197 inhibition suppresses tumour growth and has therapeutic relevance for NSCLC (Fiori et al, 2014). A previous study had also shown that miR-197 targets the tumour suppressor FUS1 (Du et al, 2009). In addition, Yanaihara et al (2006) reported, based on microarray data, that miR-197 is upregulated in cancerous versus non-cancerous lung tissues. In our study, we did not observe such an upregulation; this might be attributed to the limited sample size of non-cancerous tissues that we had available and/or the different techniques for miR-197 determination that were used between the two studies. However, we found that tissue miR-197 expression was associated with the presence of larger tumours (P=0.042) and the squamous cell carcinoma histotype (P=0.032). MiR-197 has also been described as oncogenic in a series of other aggressive malignancies. In hepatocellular carcinoma, miR-197 is upregulated and linked with enhanced cell migration, invasion and metastasis both in vitro and in vivo (Dai et al, 2014). In pancreatic cancer, miR-197 is upregulated and has been shown to promote cell migration, invasion, epithelial–mesenchymal transition and metastasis (Hamada et al, 2013). In thyroid cancer, the observed upregulation of miR-197 has been associated with carcinogenesis (Weber et al, 2006).
Despite following the internal validation and quality control procedures, the results of our study still require rigorous external validation though independent, larger and ideally multicentric cohorts, to comprehensively corroborate the prognostic significance of miR-197. Given the previously reported biological link between miR-197 and p53, it would be interesting to investigate if the prognostic performance of miR-197 can be improved by including the p53 mutation status of NSCLC patients. Considering that the qPCR technique that we used cannot account for tissue heterogeneity, it would be fitting to analyse, in a future study, the expression of miR-197 through in situ hybridisation to evaluate if there is any association between miR-197 staining intensity in distinct tissue parts and tumour aggressiveness and/or oncologic outcome. Another interesting perspective would be the clinical evaluation of tissue or circulating miR-197 levels as a biomarker for predicting or monitoring chemotherapy response in NSCLC.
According to our data, the cost-effective expression analysis of miR-197 at the time of diagnosis could be considered further as a novel molecular tool that could contribute to a more optimised clinical management for NSCLC patients.
Change history
28 April 2015
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Abd-El-Fattah AA, Sadik NA, Shaker OG, Aboulftouh ML (2013) Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem Biophys 67 (3): 875–884.
Boeri M, Pastorino U, Sozzi G (2012) Role of microRNAs in lung cancer: microRNA signatures in cancer prognosis. Cancer J 18 (3): 268–274.
Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabro E, Croce CM, Pastorino U, Sozzi G (2011) MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA 108 (9): 3713–3718.
Camp RL, Dolled-Filhart M, Rimm DL (2004) X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 10 (21): 7252–7259.
Chen L, Li C, Peng Z, Zhao J, Gong G, Tan D (2013) miR-197 expression in peripheral blood mononuclear cells from hepatitis B virus-infected patients. Gut Liver 7 (3): 335–342.
Choi SY, Yun J, Lee OJ, Han HS, Yeo MK, Lee MA, Suh KS (2013) MicroRNA expression profiles in placenta with severe preeclampsia using a PNA-based microarray. Placenta 34 (9): 799–804.
Dai N, Zhong ZY, Cun YP, Qing Y, Chen C, Jiang P, Li MX, Wang D (2013) Alteration of the microRNA expression profile in human osteosarcoma cells transfected with APE1 siRNA. Neoplasma 60 (4): 384–394.
Dai W, Wang C, Wang F, Wang Y, Shen M, Chen K, Cheng P, Zhang Y, Yang J, Zhu R, Zhang H, Li J, Zheng Y, Lu J, Zhou Y, Xu L, Guo C (2014) Anti-miR-197 inhibits migration in HCC cells by targeting KAI 1/CD82. Biochem Biophys Res Commun 446 (2): 541–548.
Du L, Schageman JJ, Subauste MC, Saber B, Hammond SM, Prudkin L, Wistuba II, Ji L, Roth JA, Minna JD, Pertsemlidis A (2009) miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res 7 (8): 1234–1243.
Duffy MJ, Crown J (2008) A personalized approach to cancer treatment: how biomarkers can help. Clin Chem 54 (11): 1770–1779.
Estep M, Armistead D, Hossain N, Elarainy H, Goodman Z, Baranova A, Chandhoke V, Younossi ZM (2010) Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment Pharmacol Ther 32 (3): 487–497.
Fiori ME, Barbini C, Haas TL, Marroncelli N, Patrizii M, Biffoni M, De Maria R (2014) Antitumor effect of miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ 21 (5): 774–782.
Gadgeel SM, Ramalingam SS, Kalemkerian GP (2012) Treatment of lung cancer. Radiol Clin North Am 50 (5): 961–974.
Goodgame B, Viswanathan A, Miller CR, Gao F, Meyers B, Battafarano RJ, Patterson A, Cooper J, Guthrie TJ, Bradley J, Pillot G, Govindan R (2008) A clinical model to estimate recurrence risk in resected stage I non-small cell lung cancer. Am J Clin Oncol 31 (1): 22–28.
Hamada S, Satoh K, Miura S, Hirota M, Kanno A, Masamune A, Kikuta K, Kume K, Unno J, Egawa S, Motoi F, Unno M, Shimosegawa T (2013) miR-197 induces epithelial-mesenchymal transition in pancreatic cancer cells by targeting p120 catenin. J Cell Physiol 228 (6): 1255–1263.
Jernas M, Malmestrom C, Axelsson M, Nookaew I, Wadenvik H, Lycke J, Olsson B (2013) MicroRNA regulate immune pathways in T-cells in multiple sclerosis (MS). BMC Immunol 14: 32.
Johnson DH, Schiller JH, Bunn PA Jr. (2014) Recent clinical advances in lung cancer management. J Clin Oncol 32 (10): 973–982.
Jones R, Payne B (1997) Clinical Investigation and Statistics in Laboratory Medicine. ACB Venture Publications: London.
Karolina DS, Tavintharan S, Armugam A, Sepramaniam S, Pek SL, Wong MT, Lim SC, Sum CF, Jeyaseelan K (2012) Circulating miRNA profiles in patients with metabolic syndrome. J Clin Endocrinol Metab 97 (12): E2271–E2276.
Keutgen XM, Filicori F, Crowley MJ, Wang Y, Scognamiglio T, Hoda R, Buitrago D, Cooper D, Zeiger MA, Zarnegar R, Elemento O, Fahey TJ 3rd (2012) A panel of four miRNAs accurately differentiates malignant from benign indeterminate thyroid lesions on fine needle aspiration. Clin Cancer Res 18 (7): 2032–2038.
Laios A, O’Toole S, Flavin R, Martin C, Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, D’Arcy T, McGuinness E, Sheils O, Sheppard B, O’Leary J (2008) Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer 7: 35.
Lehmann U, Streichert T, Otto B, Albat C, Hasemeier B, Christgen H, Schipper E, Hille U, Kreipe HH, Langer F (2010) Identification of differentially expressed microRNAs in human male breast cancer. BMC Cancer 10: 109.
Lerman G, Avivi C, Mardoukh C, Barzilai A, Tessone A, Gradus B, Pavlotsky F, Barshack I, Polak-Charcon S, Orenstein A, Hornstein E, Sidi Y, Avni D (2011) MiRNA expression in psoriatic skin: reciprocal regulation of hsa-miR-99a and IGF-1R. PLoS One 6 (6): e20916.
Leung F, Diamandis EP, Kulasingam V (2012) From bench to bedside: discovery of ovarian cancer biomarkers using high-throughput technologies in the past decade. Biomark Med 6 (5): 613–625.
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, Sun L, Ji G, Shi Y, Han Z, Han S, Nie Y, Chen X, Zhao Q, Ding J, Wu K, Daiming F (2011) miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res 9 (7): 824–833.
Liloglou T, Bediaga NG, Brown BR, Field JK, Davies MP (2014) Epigenetic biomarkers in lung cancer. Cancer Lett 342 (2): 200–212.
Lin J, Beer DG (2012) Molecular predictors of prognosis in lung cancer. Ann Surg Oncol 19 (2): 669–676.
Liu C, Iqbal J, Teruya-Feldstein J, Shen Y, Dabrowska MJ, Dybkaer K, Lim MS, Piva R, Barreca A, Pellegrino E, Spaccarotella E, Lachel CM, Kucuk C, Jiang CS, Hu X, Bhagavathi S, Greiner TC, Weisenburger DD, Aoun P, Perkins SL, McKeithan TW, Inghirami G, Chan WC (2013) MicroRNA expression profiling identifies molecular signatures associated with anaplastic large cell lymphoma. Blood 122 (12): 2083–2092.
Maes OC, Chertkow HM, Wang E, Schipper HM (2009) MicroRNA: implications for alzheimer disease and other human CNS disorders. Curr Genomics 10 (3): 154–168.
Martini N, Bains MS, Burt ME, Zakowski MF, McCormack P, Rusch VW, Ginsberg RJ (1995) Incidence of local recurrence and second primary tumors in resected stage I lung cancer. J Thorac Cardiovasc Surg 109 (1): 120–129.
Mavridis K, Stravodimos K, Scorilas A (2013) Downregulation and prognostic performance of microRNA 224 expression in prostate cancer. Clin Chem 59 (1): 261–269.
Minguzzi S, Selcuklu SD, Spillane C, Parle-McDermott A (2014) An NTD-associated polymorphism in the 3′ UTR of MTHFD1L can affect disease risk by altering miRNA binding. Hum Mutat 35 (1): 96–104.
Nana-Sinkam SP, Croce CM (2013) Clinical applications for microRNAs in cancer. Clin Pharmacol Ther 93 (1): 98–104.
Nikiforova MN, Tseng GC, Steward D, Diorio D, Nikiforov YE (2008) MicroRNA expression profiling of thyroid tumors: biological significance and diagnostic utility. J Clin Endocrinol Metab 93 (5): 1600–1608.
Ninomiya M, Kondo Y, Funayama R, Nagashima T, Kogure T, Kakazu E, Kimura O, Ueno Y, Nakayama K, Shimosegawa T (2013) Distinct microRNAs expression profile in primary biliary cirrhosis and evaluation of miR 505-3p and miR197-3p as novel biomarkers. PLoS One 8 (6): e66086.
Pereira PM, Marques JP, Soares AR, Carreto L, Santos MA (2010) MicroRNA expression variability in human cervical tissues. PLoS One 5 (7): e11780.
Peters S, Adjei AA, Gridelli C, Reck M, Kerr K, Felip E (2012) Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 23 (Suppl 7): vii56–vii64.
Powell CA, Halmos B, Nana-Sinkam SP (2013) Update in lung cancer and mesothelioma 2012. Am J Respir Crit Care Med 188 (2): 157–166.
Qi Y, Zhu Z, Shi Z, Ge Y, Zhao K, Zhou M, Cui L (2014) Dysregulated microRNA expression in serum of non-vaccinated children with varicella. Viruses 6 (4): 1823–1836.
Reck M, Heigener DF, Mok T, Soria JC, Rabe KF (2013) Management of non-small-cell lung cancer: recent developments. Lancet 382 (9893): 709–719.
Rivas MA, Venturutti L, Huang YW, Schillaci R, Huang TH, Elizalde PV (2012) Downregulation of the tumor-suppressor miR-16 via progestin-mediated oncogenic signaling contributes to breast cancer development. Breast Cancer Res 14 (3): R77.
Shen J, Jiang F (2012) Applications of microRNAs in the diagnosis and prognosis of lung cancer. Expert Opin Med Diagn 6 (3): 197–207.
Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T (2004) Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64 (11): 3753–3756.
Vannini I, Fanini F, Fabbri M (2013) MicroRNAs as lung cancer biomarkers and key players in lung carcinogenesis. Clin Biochem 46 (10-11): 918–925.
Vansteenkiste J, De Ruysscher D, Eberhardt WE, Lim E, Senan S, Felip E, Peters S (2013) Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 24 (Suppl 6): vi89–vi98.
Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103 (7): 2257–2261.
Wang P, Li Y, Hong W, Zhen J, Ren J, Li Z, Xu A (2012) The changes of microRNA expression profiles and tyrosinase related proteins in MITF knocked down melanocytes. Mol Biosyst 8 (11): 2924–2931.
Wang T, Zhang X, Obijuru L, Laser J, Aris V, Lee P, Mittal K, Soteropoulos P, Wei JJ (2007) A micro-RNA signature associated with race, tumor size, and target gene activity in human uterine leiomyomas. Genes Chromosomes Cancer 46 (4): 336–347.
Weber F, Teresi RE, Broelsch CE, Frilling A, Eng C (2006) A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. J Clin Endocrinol Metab 91 (9): 3584–3591.
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI (2008) Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res 14 (9): 2588–2592.
Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9 (3): 189–198.
Yang C, Wang C, Chen X, Chen S, Zhang Y, Zhi F, Wang J, Li L, Zhou X, Li N, Pan H, Zhang J, Zen K, Zhang CY, Zhang C (2013a) Identification of seven serum microRNAs from a genome-wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer 132 (1): 116–127.
Yang Y, Li YX, Yang X, Jiang L, Zhou ZJ, Zhu YQ (2013b) Progress risk assessment of oral premalignant lesions with saliva miRNA analysis. BMC Cancer 13: 129.
Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, Shah A, Willeit J, Mayr M (2010) Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 107 (6): 810–817.
Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, Renard JM, Mayr A, Weger S, Schett G, Shah A, Boulanger CM, Willeit J, Chowienczyk PJ, Kiechl S, Mayr M (2012) Prospective study on circulating MicroRNAs and risk of myocardial infarction. J Am Coll Cardiol 60 (4): 290–299.
Zheng D, Haddadin S, Wang Y, Gu LQ, Perry MC, Freter CE, Wang MX (2011) Plasma microRNAs as novel biomarkers for early detection of lung cancer. Int J Clin Exp Pathol 4 (6): 575–586.
Zhou J, Zheng Q, Xu T, Liao D, Zhang Y, Yang S, Hu J (2014) Associations between physical activity-related miRNAs and metabolic syndrome. Horm Metab Res 46 (3): 201–205.
Zhou J, Zhou Y, Yin B, Hao W, Zhao L, Ju W, Bai C (2010) 5-Fluorouracil and oxaliplatin modify the expression profiles of microRNAs in human colon cancer cells in vitro. Oncol Rep 23 (1): 121–128.
Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, Thomas RK, Naoki K, Ladd-Acosta C, Liu N, Pintilie M, Der S, Seymour L, Jurisica I, Shepherd FA, Tsao MS (2010) Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol 28 (29): 4417–4424.
Acknowledgements
We thank Professor Pascal Dumont (Tours Hospital), Valérie Gissot, Lysiane Brick, Géraldine Meunier, Alexandra Fayault and Aliette Decock-Giraudaud (CIC INSERM 1415) for their help in collecting tissues and clinical data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
From twelve months after its original publication, this work is licensed under the Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Mavridis, K., Gueugnon, F., Petit-Courty, A. et al. The oncomiR miR-197 is a novel prognostic indicator for non-small cell lung cancer patients. Br J Cancer 112, 1527–1535 (2015). https://doi.org/10.1038/bjc.2015.119
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2015.119
- Springer Nature Limited
Keywords
This article is cited by
-
Prognostic value of plasma microRNAs for non-small cell lung cancer based on data mining models
BMC Cancer (2024)
-
Cancer-derived exosomal miR-197-3p confers angiogenesis via targeting TIMP2/3 in lung adenocarcinoma metastasis
Cell Death & Disease (2022)
-
Familial Mediterranean fever-related miR-197-3p targets IL1R1 gene and modulates inflammation in monocytes and synovial fibroblasts
Scientific Reports (2021)
-
Establishment of the prognostic index of lung squamous cell carcinoma based on immunogenomic landscape analysis
Cancer Cell International (2020)
-
miR-197-3p reduces epithelial–mesenchymal transition by targeting ABCA7 in ovarian cancer cells
3 Biotech (2020)