Abstract
Frost in late spring is one form of environmental stress that severely damages grapevines. Starch is a major product of photosynthesis that plays essential roles in many biological processes in plants. The dynamics of starch metabolism and related gene expression in the leaves of grapevine during chilling stress have remained unclear. Here, starch metabolism in the leaves of Vitis vinifera cv. Cabernet Sauvignon was characterized under normal (25℃) and late-spring frost mimic (4℃) conditions. The results from anthracenone colorimetry, iodine staining and ultrathin sectioning are consistent with a low temperature during the night inhibiting the degradation of starch. Four α-amylases (AMY) and ten β-amylases (BAM) genes were identified in the V. vinifera genome (PN40024). Low nighttime temperatures downregulated the expression of genes that encode amylases relative to optimal nighttime temperatures. The expression of other genes with functions related to starch degradation, such as like starch excess four 1 (LSF1) and isoamylase 3 (ISA3), was induced by low temperature. The glucose, sucrose, maltose, and fructose contents increased in plants grown under low-temperature conditions with less consumption of starch, indicating complex regulation of soluble sugars. The findings here provide clues that will lead to enhanced frost tolerance of grapevine leaves by modifying the starch degradation pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Frost in late spring is a climate phenomenon characterized by the temperature suddenly dropping below zero degrees centigrade. This phenomenon often causes freezing damage to newly germinated seedlings or buds on plants and thus decreases the yield and quality of crops (Rodrigo et al. 2000). Plants have evolved several strategies to avoid damage from chilling or freezing temperatures, such as enhanced reactive oxygen species (ROS) scavenging and osmotic regulation by accumulating soluble sugars and proteins (Koster and Lynch 1992; Renaut et al. 2004; Patton et al. 2007; Theocharis et al. 2012; Saddhe et al. 2020). Uncovering the regulatory mechanisms underpinning the low-temperature response will provide key information for improving the chilling tolerance of plants by genetic engineering.
Starch is one of the main end products of photosynthesis, accumulates in plastids and is the most widely distributed nonstructural carbohydrate in plants (Sulpice 2009; Stitt and Zeeman 2012; Streb and Zeeman 2012). As the main energy storage carbohydrate, starch plays an important role in different developmental stages and multiple stress responses in plants (Smith et al. 1993; Thalmann, et al. 2017). Starch usually accumulates as a major end product of photosynthesis during the day and is almost completely degraded into hexose and transported into sink tissues during the night (Lloyd et al. 2005). ADP-glucose pyrophosphorylase (AGPase), granule-bound starch synthase (GBSS), soluble starch synthetase (SSS), starch branching enzyme (SBE) and debranching enzyme (DBE) are key enzymes responsible for starch biosynthesis (Orzechowski 2008; Yu et al. 2015; Smith 2012). Genes that encode α-amylase (AMY), β-amylase (BAM) and debranching enzymes are responsible for the degradation of starch (Fulton et al. 2008; Kötting et al. 2005; Streb and Zeeman, 2012).
The processes of starch accumulation and degradation are modified under stress conditions to enhance tolerance to harsh environments (Thalmann et al. 2016; Zanella et al. 2016; Zhu et al. 2021). Previous studies have demonstrated essential roles for starch in the supply of metabolites and energy required to tolerate cold stress in plants. Maltose, the main product of starch breakdown, increased rapidly when Arabidopsis thaliana was subjected to low temperature stress (Kaplan and Guy et al. 2004). Starch is hydrolyzed to osmotic substances, such as hexose, which increases cell fluid concentrations to help cope with low-temperature stress (Savitch, 1997; Sicher et al. 2011; Sitnicka et al. 2014; Dong et al. 2019). Compared with AMY genes, BAM genes were found to be more closely correlated with starch degradation during stress responses in plants (Yu et al. 2005; Peng et al. 2014; Monroe et al. 2014; Purdy et al. 2013). For example, the overexpression of BAM3 in Nicotiana tabacum and Pyrus betulaefolia promoted the degradation of starch into soluble sugars as a penetrant or antioxidant to improve cold tolerance (Zhao et al. 2019). Overexpression of cold-inducible AaBAM3.1 improved the cold tolerance of kiwifruit by increasing the soluble sugar content (Sun et al. 2021). These results indicate the potential utilization of BAM genes as key regulators for improving the cold tolerance of plants by genetic engineering.
Grapevine is cultivated in many countries. Newly germinated leaves, branches and inflorescences of grapevine often suffer from freezing stress during late-spring frost, which causes severe damage to grapevines and the vine industry. Previous studies have indicated that the expression of genes associated with starch pathways changes in response to chilling stress in grapevine. For example, the expression of an AMY gene was induced after chilling treatment in the daytime in leaves of Vitis vinifera (V. vinifera) and V. amurensis (Xin et al. 2013; Xu et al. 2014). The expression of five BAM genes was increased in grapevine leaves after 24 h and 72 h of chilling treatment at 4℃ (Chai et al. 2019). Overexpression of VvBAM1 improved the cold hardiness of transgenic tomato by increasing the soluble sugar content and enhancing the ROS scavenging capacity (Liang et al. 2021). Nonetheless, the dynamics of starch metabolism and the expression of associated pathway genes in grapevine leaves in response to sudden exposure to low temperatures at night are still unknown.
In this study, the starch content of leaves from potted V. vinifera cv. Cabernet Sauvignon seedlings that were grown in optimal and frost-mimic conditions were quantified using different methods. The AMY genes in the grapevine (V. vinifera cv. Pinot noir PN40024) genome were identified, and AMY expression under chilling stress conditions was quantified. The dynamics of starch degradation products, such as glucose and maltose, and the expression of genes that encode enzymes related to these processes were also quantified. The results provide new clues for strategies that might enhance the cold tolerance of grapevine leaves during late spring frosts by regulating starch degradation.
Materials and methods
Plant materials and chilling treatment
One-year-old grafted seedlings of V. vinifera cv. Cabernet Sauvignon were grown in 5-L pots containing peat soil:vermiculite (1:1) at 26°C with 12 h of light (from 6:00 to 18:00) and 12 h of dark (18:00 to 6:00) in a greenhouse. Seedlings with 5–6 well-developed leaves were placed in an illuminated incubation chamber with a photoperiod containing 12 h of light. After 3 d, the third to fourth fully developed leaves were collected at 6:00, 12:00 and 18:00 at 25℃. Then, the temperature was reduced to 4°C to mimic frost conditions in the dark within 30 min, and the leaves were collected at 18:00, 21:00, 24:00, 3:00 and 6:00. Leaf samples grown under optimal conditions (25℃) were collected at the same time and served as controls. Three biological replicates were collected at each time point. Each biological replicate came from three plantlets. The collected samples were fixed with 2.5% glutaraldehyde suitable for electron microscopy. For iodine staining experiments and observation of ultrastructure and for gene expression, starch and sugar content analysis, tissue was suddenly frozen in liquid nitrogen and then stored at -80°C until it was analyzed.
Identification of the AMY gene families in grapevine
The amino acid sequences of AMY and BAM proteins from Arabidopsis thaliana (A. thaliana) and Oryza sativa were obtained from the TAIR database (https://www.arabidopsis.org/) and RAP-DB (https://rapdb.dna.affrc.go.jp/), respectively. These amino acid sequences were used as queries in blastp searches for candidate AMY genes in the grape genome (https://phytozome-next.jgi.doe.gov/) with a cutoff e-value < 10–5. To further identify all AMY and BAM genes in grapevine, NCBI CD searches and Pfam were used to identify the conserved domains in AMY (PF07821 and PF00128) and BAM (PF01373). Proteins containing conserved domains were retained. The amylase genes in grapevine were named based on the established amylase gene nomenclature in A. thaliana. Expasy-ProtParam (https://web.expasy.org/protparam/) was used to predict the chemical and physical properties of the amylase proteins. The location of these genes was obtained from genome annotation files (http://genomes.cribi.utnipd.it/DATA/). TargetP-2.0 (https://services.healthtech.dtu.dk/service.php?TargetP-2.0), Chlorop1.1 (http://www.cbs.dtu.dk/services/ChloroP/) and WoLF PSORT (https://wolfpsort.hgc.jp/) were used to predict the subcellular localization of the amylase proteins.
All amylase amino acid sequences from grapevine, rice, and A. thaliana were aligned using ClustalW with the default settings. The neighbor-joining method in MEGA7.0 software was used to construct an unrooted phylogenetic tree with a bootstrap value of 1,000. All sequences containing the core functional domains of AMY and BAM were aligned using the ClustalX program with default settings followed by visualization with CLC Sequence Viewer6.0. The conserved motifs in amylase proteins were identified using Multiple Em for Motif Elicitation (MEME) (https://meme-suite.org/meme/tools/meme), a program available online.
Promoter cis-regulatory element analysis for VvAMY and VvBAM genes
Promoter regions were defined as 2,000 bp upstream of the translation start codon for each gene and were downloaded from the grapevine genome sequence (https://phytozome-next.jgi.doe.gov/). The PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to predict conserved cis elements in the promoter sequences.
RNA isolation and quantitative real-time PCR (qRT‒PCR)
The RNAprep Pure Plant Plus Kit (Tiangen, Beijing, China) was used to extract total RNA from the samples according to the manufacturer’s instructions. The concentration and integrity of the isolated total RNAs were determined using a NanoDrop ND 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and 1.2% agarose gel electrophoresis. cDNA was synthesized using TransScript One-step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). qRT‒PCR was performed using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) with StepOne Plus (ABI, USA). The primers for each gene (Table S1) were designed using Primer Preimer 5. The β-actin gene (GenBank accession NO: EC969944) was chosen as a reference gene. The relative expression of each target gene was quantified relative to the reference gene using the 2−ΔΔCT method.
Iodine staining and transmission electron microscopy (TEM)
Samples collected at each time point were stained with iodine using published protocols (Eimert et al. 1995; Wang, et al. 2014). Briefly, leaves were depigmented in 95% ethanol for 24 h and then washed twice with deionized water. Rehydrated leaves were stained in an I2-KI solution containing 8 g of iodine and 1 g of potassium iodide in 100 mL of distilled water for 10 min in the dark and then incubated in water until a clear background was obtained.
For TEM, leaves were fixed in electron microscopy fixation fluid (2.5% glutaraldehyde) overnight at 4℃. Leaves were rinsed in 0.1 M phosphate buffer, pH 7.2, three times. Each rinse was for 15 min. Samples were then fixed with 0.1 M phosphate buffer, pH 7.4, 1% osmic acid for 7 h at room temperature, followed by rinsing in 0.1 M phosphate buffer, pH 7.2, three times. Each rinse was for 15 min. Leaves were dehydrated in a series of 30%, 50%, 70%, 80%, 95%, 100%, 100% (1 h each). Tissues were penetrated in ethanol-acetone mixtures (3: 1, 0.5 h; 1: 1; 0.5 h; 1: 3, 0.5 h (v: v)) and in acetone for 1 h. Leaves were embedded in acetone-EMBed 812 mixtures (3: 1, 4 h; 1: 1, overnight; 1: 3, 4 h (v: v)) and in pure EMBed 812 for 8 h at 37℃. Finally, leaves were embedded in blocks that were incubated at 37°C overnight. Subsequently, samples were allowed to polymerize in a 65℃ oven for more than 48 h. Ultrathin Sects. (70 nm) were stained with uranyl acetate and lead citrate. TEM observations were performed on an HT7800 transmission electron microscope (Hitachi, Japan) with an accelerating voltage of 80 kV (Yandeau-Nelson et al. 2011).
Determination of starch content
Starch content was determined using the anthrone-sulfuric acid method (Magné et al. 2006). First, 0.2 g of leaves was ground with liquid nitrogen and added to 2 mL of 80% ethanol. Then, the samples were incubated in an 80°C water bath for 30 min followed by centrifugation at 3,000 g for 10 min. The supernatant was removed. Second, 2 mL of ultrapure water was added to the precipitate to facilitate gelatinization (15 min). Third, 2 mL of 9.2 mol/L HCIO4 was added to the precipitate with stirring for 15 min. Then, 4 mL of ultrapure water was added to the mixture and mixed well. After centrifugation at 3,000 g for 10 min, the supernatant was transferred to a new tube. Fourth, 4.6 mol/L HCIO4 was added to the precipitate with stirring for 15 min, and then 5 mL of ultrapure water was added to the mixture and mixed well. After centrifugation at 3,000 g for 10 min, the supernatant was mixed with the previous supernatant. Finally, the precipitate was washed twice with ultrapure water, and the supernatants were mixed with the previous supernatant. Then, 2 mL of the supernatant was mixed with 5 mL of an anthrone-sulfuric acid solution containing 0.2 g of anthrone and 1.0 g of thiourea dissolved in 100 mL concentrated sulfuric acid and incubated in 100℃ water baths for 10 min. The sample was rinsed with running water at room temperature to promote cooling. The absorbance at 620 nm was measured with a spectrophotometer (TECAN, M200 PR, Austria). The standard curve was constructed with the absorbance values of solutions containing different concentrations of glucose (0 mg/mL, 0.2 mg/mL, 0.4 mg/mL, 0.6 mg/mL, 0.8 mg/mL, 1 mg/mL) and used to calculate the concentration of starch in each sample.
Determination of soluble sugar content
The soluble sugar content was measured using high-performance liquid chromatography (HPLC, Zhao et al. 2018). A total of 0.2 g leaves was ground with a mortar and pestle and then transferred to tubes containing 2 mL of 80% ethanol. The tubes were incubated in an 80°C water bath for 30 min followed by centrifugation at 3000 g for 10 min to collect the supernatant. The precipitate was then washed twice with 80% ethanol and mixed with the previous supernatant. Then, the solution was evaporated under a stream of nitrogen and redissolved in ultrapure water. The solution was filtered through a PTFE 0.2 μm filter. HPLC was carried out using an Agilent 1290 Infinity II system (Germany) with an Agilent Polaris NH2 chromatographic column (250 × 4.6 mm, 5 μm). The flow rate was set at 0.8 mL/min, and the column temperature was 40°C. The mobile phase was acetonitrile–water (75:25, v/v). A refractive index detector was used and set at 40°C. The injection volume was 5 μL. Standard curves for glucose, fructose, sucrose, and maltose were prepared in the concentration range of 0.5–2 mg/mL with three replicates. The standard curve was plotted on the basis of sugar concentration and peak area. The slope, intercept and correlation coefficient of each curve were calculated using linear regression analysis. The concentration of each soluble sugar in the sample was calculated using the standard curve.
Results
Inhibition of starch degradation in grapevine leaves at a low temperature
In general, the rapid cooling of late-spring frosts occurs at night. To mimic late-spring frost conditions, we used a nighttime temperature of 4℃. The changes in starch content during the day (25℃) and night (both 25℃ and 4℃) were compared using both qualitative and quantitative methods. First, iodine staining was performed to observe starch in leaves. The blue color from the staining procedure increased gradually in leaves from 6:00 to 18:00, which indicated that the synthesis and accumulation of starch from photosynthesis increased during the day (Fig. 1a). The intensity of the stain diminished in the leaves that were collected during the night at 25℃, which is consistent with the almost complete degradation of starch in the leaves at night at 25℃. However, there was no obvious difference in the iodine staining of the leaves collected during the night at 4℃ relative to the leaves collected during the day at 25℃ (Fig. 1a), which indicates that the degradation of starch that occurs at 25℃ was blocked by the low temperature treatment. The ultramicroscopic structure of leaves was also examined to observe the starch grains in chloroplasts. As shown in Fig. 1b, the size of the starch grains increased dramatically during the day and then almost disappeared during the night at 25℃. In contrast, starch grains were still observable in the chloroplasts of the 4℃-treated leaves (Fig. 1b). Furthermore, we quantified the starch content using the anthrone-sulfuric acid method and found that the starch content in grapevine leaves accumulated at 25℃ during the day (6:00–18:00), reached a daily peak of 40 mg/g at 18:00 (Fig. 1c), and then declined to approximately 5 mg/g by 6:00 the following day. However, for the leaves subjected to the low temperature (4℃) treatment, the starch content was approximately 25 mg/g at 6:00 the next day. All these results demonstrate the diurnal dynamics of starch metabolism in grapevine leaves and that the frost stress mimic treatment inhibits the starch degradation that occurs during the night.
Diurnal dynamics of starch content in grapevine leaves at optimal and cold temperatures (a) Iodine-stained grapevine leaves. (b) Ultrastructure of grapevine leaves. C, chloroplast; S, starch grain. (c) Starch content of grapevine leaves. The data points represent mean values ± SEs from three replicates. ** and * indicate statistically significant differences determined using Student’s t test (P < 0.01 and P < 0.05, respectively) between the data collected at different temperatures
Identification and characterization of VvAMY and VvBAM genes in grapevine
We speculated that the reduced expression of AMY genes may explain the inhibition of starch degradation at low temperatures. We used blastp to identify the AMY and BAM genes in the V. vinifera cv. Pinot Nior PN40024 genome (Jaillon et al. 2007). The conserved domains were identified using NCBI CD and Pfam. In a previous study, six members of the BAM gene family were identified (Liang et al. 2021). Here, a total of four VvAMY genes and ten VvBAM genes were identified. Detailed information on these genes, including gene names, ORF lengths, molecular weight and subcellular localizations of encoded proteins, is summarized in Table 1. The phylogenetic relationships among AMY genes from grapevine, Arabidopsis (Stanley et al. 2002; Monroe et al. 2018) and rice (Yue et al. 2019; Koide et al. 2011) were determined (Fig. 2a). Genes from the AMY or BAM families from different species form two clades. The results from the conserved motif prediction analysis showed that the VvAMY and VvBAM sequences contain similar motifs (Fig. 2b, c). We found structural variation in two VvBAM amino acid sequences, including the absence of motifs 2, 3, and 6 in VvBAM1a and the absence of motif 9 in VvBAM8. For more information on the variation that may influence catalytic activity, the amino acid sequences of VvAMY and VvBAM proteins were clustered (Zhang and Li 2017). The catalytic sites (i.e. the Asp-Glu-Asp motif) were highly conserved in VvAMY sequences (Fig. 2d). BAMs that belong to the GH-14 family are characterized by a conserved core glucosyl hydrolase domain (Fig. 2e). In particular, glutamic acid residues Glu186 and Glu380 of soybean BAMs play essential roles as general acid and general base catalysts, respectively (Kang et al. 2004). A multisequence alignment of VvBAM amino acids revealed that Glu186 was highly conserved and that only Glu380 was not conserved in VvBAM2 and VvBAM7. Structural differences in BAM families indicate varied enzyme activities among members.
Phylogenetic and conserved motif analysis of VvAMYs and VvBAMs (a) Phylogenetic analysis of amylases from grapevine, Arabidopsis and rice. The amino acid sequences from Arabidopsis, rice, and grapevine are indicated with yellow squares, green circles and purple triangles, respectively. The accession numbers for the 15 α-amylases (AMY) and 29 β-amylases (BAM) used in the phylogenetic tree are provided (Table S1). (b) Structural analysis of VvAMY genes. (c) Structural analysis of VvBAM genes. (d) Multiple sequence alignment of VvAMY amino acid sequences. Three catalytically important residues and the putative N-glycosylation sites are indicated with open red boxes. Two carbohydrate-binding sites in the consensus sequence are highlighted with green boxes. (e) Multiple sequence alignment of VvBAM amino acid sequences. Black arrowheads and black lines indicate substrate-binding residues. Red arrowheads indicate the two catalytic residues (Glu 186 and Glu 380)
Expression of VvAMY and VvBAM genes in grapevine leaves under optimal and low temperatures during the night
We explored the expression patterns of VvAMY and VvBAM genes in leaves at night at optimal and low temperatures (Fig. 3). The expression of all genes was detectable in grapevine leaves but with different patterns. At the optimal temperature, the expression of three AMY genes (VvAMY1, VvAMY3a and VaAMY3b) and six BAM genes (VvBAM1a, VvBAM1b, VvBAM3, VvBAM4, VvBAM7 and VvBAM9) was induced at night. The expression patterns were negatively correlated with the dynamics of starch content, which provides evidence that the upregulation of AMY and BAM expression leads to starch degradation in grapevine leaves grown at optimal temperatures at night. In contrast, the increased nighttime expression for some of these genes was more or less repressed in the cold-treated leaves. Reduced AMY and BAM expression is consistent with reduced amylase catalytic activity and may partially explain the elevated starch content in grapevine leaves grown under chilling conditions.
Expression patterns of genes encoding other enzymes associated with starch degradation
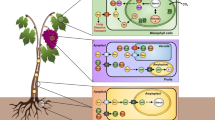
Another explanation for the elevated starch content in grapevine leaves during cold treatment is the reduced expression of other genes responsible for the degradation of starch. Other genes associated with starch degradation were identified from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.kegg.jp/pathway/map00500). Starch degradation begins with glucan water dikinase (GWD) and phosphoglucan water dikinase (PWD). GWD and PWD catalyzed the phosphorylation of starch grains, which loosened the structure of starch grains. Then, β-amylase, isoamylase (ISA) and disproportionating enzyme (DPE) degraded the starch to yield glucose and maltose. Finally, the glucose transporter (pGlcT) and maltose exporter 1 (MEX1) transport glucose and maltose across the chloroplast envelope and into the cytoplasm. We thus quantified the expression levels of these genes in leaves grown under optimal and chilling conditions. Among the eight genes we studied, the expression of only like starch excess four 1 (LSF1) and MEX1 increased during the night under optimal conditions. However, the expression of these genes was significantly upregulated in leaves subjected to chilling stress relative to leaves grown at the optimal temperature (Fig. 4). These results indicate that low temperatures induce the expression of most of the genes that encode enzymes that contribute to starch, except genes that encode amylases. Therefore, we speculate that the downregulated expression of genes that encode amylases may contribute to the reduced degradation of starch in grapevine leaves at low temperature.
Expression of genes encoding eight enzymes associated with starch catabolism under optimal and cold conditions at night. Relative expression was quantified using qRT‒PCR and calculated using the 2−△△Ct method. The relative expression of genes encoding glucan water dikinase (GWD), phosphoglucan water dikinase (PWD), like starch excess four 1 (LSF1), isoamylase 3 (ISA3), disproportionating enzyme 1 (DPE1), disproportionating enzyme 2 (DPE2), glucose transporter (pGlcT) and maltose exporter 1 (MEX1) were quantified from leaves grown under optimal and chilling conditions at night. Data are expressed as the mean values ± SDs, based on three replicates
Changes in soluble sugar content in grapevine leaves subjected to chilling treatment
Starch is one of the sources for soluble sugars that promote abiotic stress tolerance. We tested the influence of starch degradation on fructose, glucose, sucrose and maltose contents in grapevine leaves grown in normal and frost mimic conditions. The fructose and glucose contents decreased gradually under optimal conditions but remained almost unchanged during the chilling treatment (Fig. 5a, b). The sucrose content decreased at the end of the night in leaves grown at the optimal temperature but decreased first and then increased dramatically in leaves subjected to the chilling treatment (Fig. 5c). Although similar fluctuations in maltose content were found at the different temperatures, the maltose content was higher in the middle of the night in leaves grown at the chilling temperature (Fig. 5d). These results indicate that low temperatures induced the accumulation of soluble sugars, especially fructose, glucose, and sucrose. Considering that chilling treatment inhibited the degradation of starch, these data provide evidence that the regulation of soluble sugar content in grapevine leaves subjected to low-temperature stress may be complex.
Fructose a, glucose b, sucrose c and maltose d contents in grapevine leaves grown under optimal and low temperature conditions. Data are presented as the mean values ± SDs from three replicates. **and *indicate statistically significant differences relative to 0 h determined with Student’s t test (P < 0.01 and P < 0.05, respectively)
Identification of cis-regulatory elements in the promoters of VvAMY and VvBAM genes
The cis-regulatory elements in the 2.0 kb region upstream of the VvAMY and VvBAM genes were analyzed. A large number of core cis-elements are present in these genes, such as the CAAT-box and TATA-box (summarized in Fig. S1). Sequences similar to light-responsive elements were found in the promoters of all VvAMY and VvBAM genes analyzed, including Box 4, the G-box, and the GT1 motif (Fig. S1). Sequences similar to cis-elements that respond to phytohormones, including abscisic acid, auxin, salicylic acid, MeJA and gibberellin, were also identified (Fig. 6, 7). Promoter elements associated with abiotic stress, such as ABREs and STREs, were ubiquitously distributed among all VvAMY and VvBAM genes (Fig. S1). Low-temperature-responsive (LTR) elements were present mainly in VvAMY2, VvAMY3b, VvBAM1b, VvBAM7, VvBAM9 and VvBAM10. Sequences similar to promoter elements that confer cold and osmotic stress responsiveness (e.g. TC-rich repeats and the DRE core) were rarely found in these promoter regions (Fig. S1). Circadian response elements were found only in the promoter region of VvBAM8. These results provide evidence that the transcription of VvAMY and VvBAM genes is tightly related to light, phytohormone and stress responses.
Dynamics of starch degradation and related gene expression in the leaves of grapevine during chilling stress. GWD, glucan water dikinase; PWD, phosphoglucan water dikinase; LSF 1, like starch excess four 1; ISA3, isoamylase 3; AMY, α-amylases; BAM, β-amylases; DPE, disproportionating enzyme; MEX1, exporter 1; pGlct, glucose transporter
Discussion
Frost in late spring usually occurs at night, which causes severe injuries to newly geminated leaves, flowers and even the branches of fruit trees (Lamichhane 2021). Several strategies are used to reduce the damage of this meteorological disaster on fruit production, such as heaters, wind machines, overvine sprinklers and foggers (Evans 2000; Perry 2001; Tong et al. 2019). All these interventions are costly. The unpredictability of frost in late spring also limits the use of these methods. In recent years, increased knowledge of cold stress responses in plants has led to new strategies for improving the cold tolerance of plants and thus helping to overcome the damage caused by frost.
Starch is a fundamental substance in plants (Yelenosky and Guy 1977). Consistent with previous reports, we found that starch accumulates during the day and at optimal temperatures and degrades during the night in grapevine leaves (Santelia 2015; Skeffington 2014) (Fig. 1). Generally, plants remobilize starch to provide energy and carbon during stressful conditions (Thalmann 2017). Since frost in late spring always occurs at night, we designed experimental conditions that mimic this stress condition. We thought that chilling stress might accelerate the degradation of starch and therefore induce increases in the accumulation of the products of starch degradation, such as soluble sugar, to increase the energy supply and osmotic potential and thus to promote chilling tolerance. Instead, we found that the degradation of starch was dramatically blocked within a 12 h chilling treatment in the dark. Several reports have mentioned that the overexpression of amylase increases the content of soluble sugar and chilling tolerance in plants. For example, kiwifruit lines that overexpress AaBAM3.1 are more freezing tolerant than control lines (Sun et al. 2021). Thus, enhancing starch degradation should improve the chilling tolerance of grapevine leaves.
There are several AMY and BAM genes in the grapevine genome. Thus, choosing a suitable candidate for the genetic improvement of grapevine is a challenge. Among the AMY and BAM genes in grapevine, the induced expression of VvAMY genes in the dark or in response to chilling stress was weaker than we observed for the VvBAM genes. Previous work in other species indicates that the expression of AMY genes is induced by heat, drought and biotic stress and mainly affects bud dormancy, leaf senescence and seed germination (Satoru et al. 2005; Doyle et al. 2007; Rubio et al. 2014; Fan et al. 2018). BAM genes were previously found to play major roles in stress responses in plants (Peng et al. 2004; Zhao et al. 2019; Zhu et al. 2021). We found that the expression of VvBAM3 was gradually induced by chilling treatment, which is consistent with its role in starch degradation at low temperatures, and thus, VvBAM3 can be considered a candidate for the genetic improvement of grapevine. Glu380 serves a critical function as a general base catalyst and was changed in the amino acid sequences derived from VvBAM2 and VvBAM7, but the effect on enzyme activity is unknown. More work is needed to evaluate the potential of each VvBAM gene for the enhancement of cold tolerance in grapevine leaves.
Promoters are also potential targets for modifying gene expression by genetic engineering (Mao et al. 2019). Light-responsive elements are abundant in VvAMY and VvBAM gene promoter regions, which is consistent with the dark-induced expression patterns for most VvAMY and VvBAM genes. Although cis-elements associated with abiotic stress, such as LTR, ABRE, TC-rich repeats, DRE core and STRE, were identified in the promoters of VvAMY and VvBAM genes, large increases in the expression of these genes were not observed within 12 h of chilling stress. The timely response of amylase genes to cold stress might be a strategy for enhancing the soluble sugar content and thus for increasing cold tolerance when temperatures suddenly drop (Steponkus 1984). In recent years, advanced technology for gene editing has allowed for the introduction of point mutations in nucleic acids, and thus, in theory, the modification of regulated gene expression by the limited modification of promoter elements is now possible (Dietrich et al. 1996; Jang et al. 2003; Jeong et al. 2010). Further studies on the core cis-elements required for the cold-inducible expression of VvAMY and VvBAM genes may facilitate the discovery of new cultivars for grapevine and for other crops with enhanced cold tolerance without influencing growth and development (Fig. 7).
Conclusions
In conclusion, the findings of this study indicate that low temperature inhibits the degradation of starch in grapevine leaves. The expression of VvAMY and VvBAM genes at night at the optimal temperature was also inhibited by low temperature. However, the expression of other genes related to starch degradation, such as LSF1 and ISA3, was induced by low-temperature treatment. Therefore, we speculate that transcriptional regulation of genes that encode amylase may be the main factor promoting starch degradation in grapevine leaves at low temperatures. The findings presented here provide new clues that will be useful for developing strategies for enhancing the frost tolerance of grapevine leaves by modifying the starch degradation pathway.
Availability of data and materials
Data will be made available on request. All plant materials were provided free of charge. The laboratory experiments were conducted under permissions of regulations and laws.
Abbreviations
- AMY:
-
α-Amylases
- BAM:
-
β-Amylases
- LSF1:
-
Like starch excess four 1
- ISA3:
-
Isoamylase 3
- ROS:
-
Reactive oxygen species
- AGPase:
-
ADP-glucose pyrophosphorylase
- GBSS:
-
Granule-bound starch synthase
- SSS:
-
Soluble starch synthetase
- SBE:
-
Starch branching enzyme
- DBE:
-
Debranching enzyme
- TEM:
-
Transmission electron microscope
- GWD:
-
Glucan water dikinase
- PWD:
-
Phosphoglucan water dikinase
- pGlcT:
-
Glucose transporter
- MEX1:
-
Maltose exporter 1
- LTR:
-
Low temperature responsiveness
References
Chai F, Liu W, Xiang Y, Meng X, Sun X, Cheng C, Liu G, Duan L, Xin H, Li S. Comparative metabolic profiling of Vitis amurensis and Vitis vinifera during cold acclimation. Hortic Res. 2019;6:8.
Dietrich A, Maréchal-Drouard L, Carneiro V, Cosset A, Small I. A single base change prevents import of cytosolic tRNAAla into mitochondria in transgenic plants. Plant J. 1996;10(5):913–8. https://doi.org/10.1046/j.1365-313x.1996.10050913.x.
Dong S, Beckles DM. Dynamic changes in the starch-sugar interconversion within plant source and sink tissues promote a better abiotic stress response. J Plant Physiol. 2019;234–235:80–93. https://doi.org/10.1016/j.jplph.2019.01.007.
Doyle EA, Lane AM, Sides JM, Mudgett MB, Monroe JD. An alpha-amylase (at4g25000) in arabidopsis leaves is secreted and induced by biotic and abiotic stress. Plant Cell Environ. 2007;30(4):388–98.
Eimert K, Wang SM, Lue WI, Chen J. Monogenic recessive mutations causing both late floral initiation and excess starch accumulation in arabidopsis. Plant Cell. 1995;7(10):1703–12.
Evans RG. The art of protecting grapevines from low temperature injury. Proc. ASEV 50th Anniversary Annual Mtg. 2000;19–23:60–72. https://www.ars.usda.gov/ARSUserFiles/21563/Art.
Fan ZQ, Ba LJ, Shan W, Xiao YY, Lu WJ, Kuang JF, Chen JY. A banana R2R3-MYB transcription factor mamyb3 is involved in fruit ripening through modulation of starch degradation by repressing starch degradation-related genes and mabhlh6. Plant J. 2018;96(6):1191–205.
Fulton DC, Stettler M, Mettler T, Vaughan CK, Li J, Francisco PB, Gil MH, Reinhold H, Eicke S, Messerli G, Dorken G, Halliday KJ, Smith A, Smith S, Zeeman SC. β-amylase4, a noncatalytic protein required for starch breakdown, acts upstream of three active β-amylases in arabidopsis chloroplasts. The Plant Cell. 2008;20:1040–58.
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyère C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pè ME, Valle G, Morgante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quétier F, Wincker P; French-Italian Public Consortium for Grapevine Genome Characterization. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449(7161):463–7.
Jang IC, Oh SJ, Seo JS, Choi WB, Song SI, Kim CH, et al. Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth. Plant Physiol. 2003;131(2):516–24. https://doi.org/10.1104/pp.007237.
Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, et al. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153(1):185–97. https://doi.org/10.1104/pp.110.154773.
Kang YN, Adachi M, Utsumi S, Mikami B. The roles of Glu186 and Glu380 in the catalytic reaction of soybean β-amylase. J Mol Biol. 2004;339(5):1129–40. https://doi.org/10.1016/j.jmb.2004.04.029.
Kaplan F, Guy CL. β-amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 2004;135(3):1674–84. https://doi.org/10.1104/pp.104.040808.
Koide T, Ohnishi Y, Horinouchi S. Characterization of recombinant β-amylases from Oryza sativa. Biosci Biotech Bioch. 2011;75(4):793–6. https://doi.org/10.1271/bbb.100872.
Koster KL, Lynch DV. Solute accumulation and compartmentation during the cold acclimation of Puma Rye. Plant Physiol. 1992;98(1):108–13. https://doi.org/10.1104/pp.98.1.108.
Kötting O, Santelia D, Edner C, Eicke S, Marthaler T, Gentry MS, et al. STARCH-EXCESS4 is a laforin-like phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell. 2005;21(1):334–46. https://doi.org/10.1105/tpc.108.064360.
Lamichhane JR. Rising risks of late-spring frosts in a changing climate. Nat Clim Change. 2021;11(7):554–5. https://doi.org/10.1038/s41558-021-01090-x.
Liang G, He H, Nai G, Feng L, Li Y, Zhou Q, et al. Genome-wide identification of BAM genes in grapevine (Vitis vinifera L.) and ectopic expression of VvBAM1 modulating soluble sugar levels to improve low-temperature tolerance in tomato. BMC Plant Biol. 2021;21(1):156.
Lloyd JR, Kossmann J, Ritte G. Leaf starch degradation comes out of the shadows. Trends Plant Sci. 2005;10(3):130–7. https://doi.org/10.1016/j.tplants.2005.01.001.
Magné C, Saladin G, Clément C. Transient effect of the herbicide flazasulfuron on carbohydrate physiology in Vitis vinifera L. Chemosphere. 2006;62(4):650–7. https://doi.org/10.1016/j.chemosphere.2005.04.119.
Mao Y, Botella JR, Liu Y, Zhu JK. Gene editing in plants: progress and challenges. Natl Sci Rev. 2019;6(3):421–37. https://doi.org/10.1093/nsr/nwz005.
Monroe JD, Pope LE, Breault JS, Berndsen CE, Storm AR. Quaternary structure, salt sensitivity, and allosteric regulation of β-AMYLASE2 from Arabidopsis thaliana. Front Plant Sci. 2018;9:1176. https://doi.org/10.3389/fpls.2018.01176.
Monroe JD, Storm AR, Badley EM, Lehman MD, Platt SM, Saunders LK, Schmitz JM, Torres CE. β-amylase1 and β-amylase3 are plastidic starch hydrolases in arabidopsis that seem to be adapted for different thermal, pH, and stress conditions. Plant Physiol. 2014;166(4):1748–63.
Orzechowski S. Starch metabolism in leaves. Acta Biochim Pol. 2008;55(3):435–45. https://doi.org/10.1007/0-306-48137-5_9.
Patton AJ, Cunningham SM, Volenec JJ, Reicher ZJ. Differences in freeze tolerance of zoysiagrasses: II. Carbohydrate and proline accumulation. Crop Sci. 2007;47(5):2170. https://doi.org/10.2135/cropsci2006.12.0784.
Peng S, Huang J, Sheehy JE, Laza RC, Visperas RM, Zhong X, et al. Rice yields decline with higher night temperature from global warming. Proc Natl Acad Sci U S A. 2004;101(27):9971–5.
Peng T, Zhu X, Duan N, Liu JH. PtrBAM1, a β-amylase-coding gene of Poncirus trifoliata, is a cbf regulon member with function in cold tolerance by modulating soluble sugar levels. Plant Cell Environ. 2014;37(12):2754–67.
Perry KB. Frost/freeze protection for horticultural crops. Raleigh: NC State University; 2001.
Purdy SJ, Bussell JD, Nunn CP, Smith SM. Leaves of the arabidopsis maltose exporter1 mutant exhibit a metabolic profile with features of cold acclimation in the warm. PLoS One. 2013;8(11):e79412.
Renaut J, Lutts S, Hoffmann L, Hausman JF. Responses of poplar to chilling temperatures: proteomic and physiological aspects. Plant Biol (stuttg). 2004;6(1):81–90. https://doi.org/10.1055/s-2004-815733.
Rodrigo J. Spring frosts in deciduous fruit trees-morphological damage and flower hardiness. Sci Hortic. 2000;92(3):155–73.
Rubio S, Donoso A, Pérez FJ. The dormancy-breaking stimuli "chilling, hypoxia and cyanamide exposure" up-regulate the expression of α-amylase genes in grapevine buds. J Plant Physiol. 2014;171(6):373–81.
Saddhe AA, Manuka R, Penna S. Plant sugars: homeostasis and transport under abiotic stress in plants. Physiol Plant. 2021;171(4):739–55. https://doi.org/10.1111/ppl.13283.
Santelia D, Trost P, Sparla F. New insights into redox control of starch degradation. Curr Opin Plant Biol. 2015;25:1–9. https://doi.org/10.1016/j.pbi.2015.04.003.
Satoru A, Chihoko S, Kimiko I, Mitsutoshi O, Aya K, Toshiaki M. Involvement of α-amylase i–1 in starch degradation in rice chloroplasts. Plant Cell Physiol. 2005;6:858–69.
Savitch LV, Gray GR, Huner NPA. Feedback-limited photosynthesis and regulation of sucrose-starch accumulation during cold acclimation and low-temperature stress in a spring and winter wheat. Planta. 1997;201:18–26. https://doi.org/10.1007/BF01258676.
Sicher R. Carbon partitioning and the impact of starch deficiency on the initial response of Arabidopsis to chilling temperatures. Plant Sci. 2011;181(2):167–76.
Sitnicka D, Orzechowski S. Cold-induced starch degradation in potato leaves-intercultivar differences in the gene expression and activity of key enzymes. Biol Plant. 2014;58:659–66.
Skeffington AW, Graf A, Duxbury Z, Gruissem W, Smith AM. Glucan, water dikinase exerts little control over starch degradation in Arabidopsis leaves at night. Plant Physiol. 2014;165(2):866–79. https://doi.org/10.1104/pp.114.237016.
Smith A, Martin C. Starch biosynthesis and the potential for its manipulation. In: biosynthesis and manipulation of plant products. Plant Biotechnology Series. 1993. Springer, p. 54. https://doi.org/10.1007/978-94-011-2142-2_1.
Smith AM. Starch in the Arabidopsis Plant. Starch Starke. 2012;64:421–34. https://doi.org/10.1002/star.201100163.
Stanley D, Fitzgerald AM, Farnden KJF, Macrae EA, Macrae KJF. Characterisation of putative α-amylases from apple (Malus domestica) and Arabidopsis thaliana. Biologia. 2002;57(2):137–48. https://doi.org/10.1016/j.socscimed.2005.05.028.
Steponkus PL. Role of the plasma membrane in freezing injury and cold acclimation. Ann Rev Plant Physiol. 1984;35(1):543–84.
Stitt M, Zeeman SC. Starch turnover: pathways, regulation and role in growth. Curr Opin Plant Biol. 2012;15(3):282–92. https://doi.org/10.1016/j.pbi.2012.03.016.
Streb S, Zeeman SC. Starch metabolism in Arabidopsis. Arabidopsis Book. 2012;10:e0160.
Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, Von Korff M, Steinhauser MC, Keurentjes JJ, Guenther M, Hoehne M, Selbig J, Fernie AR, Altmann T, Stitt M. Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci U S A. 2009;106(25):10348–53.
Sun S, Hu C, Qi X, Chen J, Fang J. The AaCBF4-AaBAM3.1 module enhances freezing tolerance of kiwifruit (Actinidia arguta). Hortic Res. 2021;8(1):97. https://doi.org/10.1038/s41438-021-00530-1.
Thalmann M, Santelia D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017;214(3):943–51. https://doi.org/10.1111/nph.14491.
Thalmann M, Pazmino D, Seung D, Horrer D, Nigro A, Meier T. Regulation of leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell. 2016;28(8):1860–78. https://doi.org/10.1105/tpc.16.00143.
Theocharis A, Clément C, Barka EA. Physiological and molecular changes in plants grown at low temperatures. Planta. 2012;235(6):1091–105.
Tong RP, Gao YL, Liu ML, Zhai H, Sun QH. Spraying outer source of melatonin has the effect of alleviating grape leaves and frost damage. Falling Leaves. 2019;51(02):12–5.
Wang L, Zhu W, Fang L, Sun X, Su L, Liang Z. Genome-wide identification of WRKY family genes and their response to cold stress in Vitis vinifera. BMC Plant Biol. 2014;14:103. https://doi.org/10.1186/1471-2229-14-103.
Xin H, Zhu W, Wang L, Xiang Y, Fang L, Li J. Genome wide transcriptional profile analysis of Vitis amurensis and Vitis vinifera in response to cold stress. PLoS One. 2013;8(3):e58740.
Xu W, Li R, Zhang N, Ma F, Jiao Y, Wang Z. Transcriptome profiling of Vitis amurensis, an extremely cold-tolerant Chinese wild Vitis species, reveals candidate genes and events that potentially connected to cold stress. Plant Mol Biol. 2014;86(4–5):527–41. https://doi.org/10.1007/s11103-014-0245-2.
Yandeau-Nelson MD, Laurens L, Shi Z, Xia H, Smith AM, Guiltinan MJ. Starch-branching enzyme II a is required for proper diurnal cycling of starch in leaves of maize. Plant Physiol. 2011;156(2):479–90. https://doi.org/10.1104/pp.111.174094.
Yelenosky G, Guy C. Carbohydrate accumulation in leaves and stems of ‘Valencia’ orange at progressively colder temperatures. Bot Gaz. 1977;138:13–7.
Yu T, Zeeman S, Smith SM. α-Amylase is not required for breakdown of transitory starch in Arabidopsis leaves. J Biol Chem. 2005;280(11):9773–9. https://doi.org/10.1074/jbc.M413638200.
Yu X, Li B, Wang L, Chen X, Wang W, Zhong W. Systematic analysis of pericarp starch accumulation and degradation during wheat caryopsis development. PLoS One. 2015;10(9):e0138228.
Yue C, Cao H, Lin H, Hu J, Ye Y, Li J, Hao Z, Hao X, Sun Y, Yang Y, Wang X. Expression patterns of alpha-amylase and beta-amylase genes provide insights into the molecular mechanisms underlying the responses of tea plants (camellia sinensis) to stress and postharvest processing treatments. Planta. 2019;250(1):281–98.
Zanella M, Borghi GL, Pirone C, Thalmann M, Pazmino D, Costa A, Santelia D, Trost P, Sparla F. β-amylase 1 (BAM1) degrades transitory starch to sustain proline biosynthesis during drought stress. J Exp Bot. 2016;67(6):1819–26.
Zhang Q, Li C. Comparisons of copy number, genomic structure, and conserved motifs for α-Amylase genes from barley, rice, and wheat. Front Plant Sci. 2017;8:1727. https://doi.org/10.3389/fpls.2017.01727.
Zhao D, You Y, Fan H, Zhu X, Wang Y, Duan Y, et al. The role of sugar transporter genes during early infection by root-knot nematodes. Int J Mol Sci. 2018;19(1):302. https://doi.org/10.3390/ijms19010302.
Zhao LY, Yang TY, Xing CH, Dong HZ, Qi KJ, Gao JZ, et al. The β-amylase PbrBAM3 from pear (Pyrus betulaefolia) regulates soluble sugar accumulation and ROS homeostasis in response to cold stress. Plant Sci. 2019;287:110184.
Zhu H, Yang X, Wang X, Li Q, Guo J, Ma T. The sweetpotato β-amylase gene IbBAM1.1 enhances drought and salt stress resistance by regulating ROS homeostasis and osmotic balance. Plant Physiol Bioch. 2021;168:167–76. https://doi.org/10.1016/j.plaphy.2021.09.034.
Acknowledgements
This work was supported by the Grape Breeding Project of Ningxia (NXNYYZ20210104) and the Tianshan Innovation Team Project in 2021 (2020d14033).
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
HPX designed the research, PGF and LZG provided the study materials, HMZ and YJH conducted the experiments, QYL analyzed the data, DK helped to revise the manuscript, HPX and HMZ wrote the manuscript. All of the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests. The authors declare that they have no financial or nonfinancial interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, H., Li, Q., Gichuki, D.K. et al. Dynamics of starch degradation and expression of related genes during chilling stress in grapevine. HORTIC. ADV. 1, 3 (2023). https://doi.org/10.1007/s44281-023-00004-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44281-023-00004-5