Abstract
Breast cancer has been one of the leading causes of death among women in the world. Early detection of this disease can save patient’s lives and reduce mortality. Due to the large number of features involved in the diagnosis of this disease, the breast cancer diagnosis process can be time consuming. To reduce cost and time and improving accuracy of breast cancer diagnosis, this paper propose a feature selection algorithm based on particle swarm optimization (PSO) combined with machine learning methods for selection the most effective features for breast cancer diagnosis among all features. In order to evaluate the efficiency of the proposed feature selection method, it was tested on three most common breast cancer datasets available in the University of California, Irvine (UCI) repository named: Coimbra dataset (CD), Wisconsin Diagnostic Breast Cancer dataset (WDBC) and Wisconsin Prognostic Breast Cancer dataset (WPBC). In the Coimbra dataset with all its 9 features and without PSO feature selection algorithm the highest obtained accuracy was 87% by Support Vector Machine method, while with PSO feature selection algorithm the accuracy reached to 91% and the number of features was reduced from 9 to 4. In the WDBC dataset with all its 30 features and without PSO feature selection algorithm the highest obtained accuracy was 99% by Random Forest method, while with PSO feature selection algorithm the accuracy reached to 100% and the number of features was reduced from 30 to 19. In the WPBC dataset with all its 33 features and without PSO feature selection algorithm the highest obtained accuracy was 94% by Support Vector Machine method, while with PSO feature selection algorithm the accuracy reached to 96% and the number of features was reduced from 33 to 17. The results of this paper indicated that the proposed feature selection algorithm based on PSO algorithm can improve the accuracy of breast cancer diagnosis. While it has selected fewer and more effective features than the total number of features in the original datasets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Today, the use of machine learning methods to diagnose diseases have become widespread [1]. Breast cancer disease is one of the most common types of malignant cancers among worldwide women and accounts for 25.1% of all cancers [2]. Breast cancer spreads to other organs over time. Research also showed that breast cancer is more common in women whose average age is 47 years than in women whose average age is 63 years [3, 4].
Cancerous tumors are divided into malignant and benign. Benign tumors are non-intense. But malignant tumors are intensive and can spread to other parts of the body. Therefore, the correct diagnosis of the tumor for treatment must be considered. Recurrence of breast cancer can occur 1–20 years after treatment for primary cancer. Cancer patients often face treatment complications. The recurrence of breast cancer can be predicted by examining various factors such as the size of the primary tumor, the number of damaged lymph nodes, the area of the tumor, and similar factors. In recent years Machine learning models have been used in medicine to diagnose cancer and accurately classify benign and malignant tumors in a reasonable time [5, 6].
With the advancement of technology, different types of data are produced with high dimensions. The data produced in the field of medicine or cancer have wide dimensions and variables. When the dimension of the data is high, the classification results may have more error and make data analysis difficult. Also, high-dimensional data has challenges such as search space, time, and computational costs [7]. Nowadays, Machine learning in diagnosing diseases such as COVID-19 [1], cardiovascular disease, diabetes mellitus and analyzing their data was successful. Using the dimension reduction and feature selection (FS) methods makes the disease diagnosis faster, easier and less expensive. It should be easier to store and classify [8] because feature selection can produce fewer features and reduce computational costs [9]. In feature selection, the goal is to reduce the number of features in the dataset and select the most effective features so that the highest possible classification accuracy can be achieved with the least possible number of features. Machine learning methods are widely used in medical studies and automatic diagnosis of cancers such as breast cancer. Many successful detections and prediction methods have been performed, especially in studies using Coimbra, Wisconsin Diagnostic Breast Cancer (WDBC), and Wisconsin Prognostic Breast Cancer dataset (WPBC) datasets. These predictions are made using the dimensions and other features of tumors [10].
The innovation of this research is the combination of ten different machine learning algorithms including ensemble learning methods with the PSO feature selection algorithm to select the most effective features in the diagnosis of breast cancer which is implemented on three famous datasets in the field of breast cancer named Coimbra, WPBC and WDBC datasets. Also, use the PSO algorithm as a method to select more effective features in disease diagnosis and reduce the size of the dataset and by applying it to the Coimbra, WDBC and WPBC datasets to diagnose breast cancer using the most common machine learning methods and performance analysis of each algorithm. These machine learning methods include AdaBoost (ADB), Decision Tree (DT), k Nearest Neighbors (KNN), Linear Discriminant Analysis (LDA), Logistic Regression (LGR), Linear Regression (LR), Naïve Bayes (NB), Artificial Neural Networks (ANN), Random Forest (RF) and Support Vector Machine (SVM). Also, a comparative analysis between the performance evaluation criteria of machine learning methods on the original dataset and the dataset consist of the selected features by PSO feature selection algorithm was performed.
The following sections of this article are organized as follows. The second part of the article examines related works. This research includes studies on breast cancer, the use of machine learning algorithms and different techniques to diagnose breast cancer and compare their accuracy. In the third part of the article, the information related to the Coimbra, WDBC and WPBC datasets are explained, which are used to evaluate the proposed method. Also, in this section, machine learning methods such as classification methods and PSO algorithm are explained. The fourth part describes the theory and calculation of the proposed method. In the fifth section, the results obtained in this research are stated. It belongs to Discussion in the sixth part. The seventh section provides conclusions and suggestions for future research.
2 Related Works
Meta-heuristics algorithms are widely used in feature selection because they are highly efficient, easy to implement, and can manage large-scale data. Swarm Intelligence (SI) algorithm is a branch of meta-heuristics algorithms. These algorithms imitate from social behavior of animal group life, for example, Insects (instance ant, bee, etc.) birds, and fishes [11, 12]. On the other hand, feature selection is an important and challenging work in machine learning and one of its goals is to maximize the accuracy of classification [12]. For instance, in [13], the authors used the SVM methods to diagnose breast cancer and the kNN, NB, and DT algorithms to detect the type of cancer cells. In [14] a hybrid model based on concepts of neural networks and fuzzy systems presented. This model could manipulate data collected in medical examinations and detect patterns in healthy individuals and individuals with breast cancer with an acceptable level of accuracy. These intelligent techniques have made it possible to create expert systems based on logical rules of the IF/THEN type. According to its results, the hybrid model has a good capacity to predict breast cancer and analyze the characteristics of this cancer. In [10] the DT method was used to diagnose breast cancer. In this study, parameters related to blood analysis have been used. In this methods, the level of importance of the properties is determined by the Gini coefficient. Accuracy in this study is 90.5%. In [15] an analytical evaluation was performed on machine learning methods and breast cancer datasets. Some of the initial processing was performed using WEKA software on the input datasets and its overall effect on the prediction accuracy was also determined. In this research, the filter feature selection method has been used. The results show that correct feature selection can be used to select the best features and the prediction speed and accuracy can be increased. RF had the best accuracy of 69% before using the filter method and 98% after using it. Similarly, LR came in second with 96% accuracy after using the filter and 68% unfiltered, followed by NB with 91% after using the filter method and 71% unfiltered. The authors in [16] diagnosed breast cancer using four factors of resistance, glucose, age and BMI. They using three machine learning methods including SVM, RF, and LGR. They used the Monte Carlo cross-validation method to evaluate the results and the 95% confidence level. Their results indicate the superiority of the SVM method over the other two methods. In [17], four methods including Extreme Learning Machine (ELM), SVM, kNN and ANN were used to diagnose breast cancer using the Wisconsin dataset which includes blood analysis data of patients and healthy individuals. The accuracy obtained by ELM is 80%, 79.4% by the ANN method, 77.5% by the kNN method and 73.5% by the SVM method. In [18] the authors presented a SVM-based ensemble learning method that was applied on two breast cancer datasets including the Wisconsin dataset and one breast cancer dataset registered in the United States. The results of this method showed 33.34% increase in accuracy of diagnosis compared to the best individual SVM method.
3 Materials and Methods
This section examines datasets, machine learning methods, and the PSO algorithm used to increase the accuracy of breast cancer diagnosis. In this research, the data sets available in the database of the University of California Irvine (UCI), USA has been used. The UCI Machine Learning Repository is a collection of databases, domain theories, and data generators that are used by the machine learning community for the empirical analysis of machine learning algorithms. The archive was created as an ftp archive in 1987 by David Aha and fellow graduate students at UC Irvine [19].
3.1 Description of Datasets
The datasets used in this study were Coimbra, WDBC, and WPBC. The number of samples and features of which are also shown in Table 1.
3.1.1 Coimbra Dataset
The Coimbra dataset is one of the datasets used in this article that contains 116 samples. This dataset is related to a study that was performed on the obstetrics and gynecology department of Coimbra University Hospital between 2009 and 2013 and its data were collected. People have been diagnosed with breast cancer based on mammography results and through the diagnosis of specialist doctors. These data were obtained before treatment. There are ten attributes in this dataset. Among these ten attributes, one of the attributes is binary and nine attributes are continuous. The binary attribute indicates the presence or absence of cancer. Attributes are anthropometric parameters and data obtained from blood analysis [20]. The attributes and statistical parameters of the Coimbra dataset are shown in Table 2.
3.1.2 WDBC Dataset
Another dataset is WDBC. This dataset has 569 samples. Each instance contains 30 attributes in which the first attribute specifies a unique identification number and the second attribute identifies labels (357 Benign/212 Malignant). For each diagnostic sample, 30 features were computed from a digitized image of a Fine Needle Aspirate (FNA) test. Ten real valued attributes that are shown in Table 3 were computed for each cell nucleus. For each image, the mean, standard error and largest values (worst value) of these ten features were computed and giving 30 features [8, 16, 21, 22]. The ten real value attributes and statistical parameters of the WDBC dataset are shown in Table 3.
This dataset has been collected by Dr.Wolberg from patients since 1984 and there is no evidence of metastasis to other parts of the body in these patients [23].
3.1.3 WPBC Dataset
The WPBC dataset is another dataset used in this study. This dataset contains 198 samples (151 Non recurrence/47 Recurrence). Each sample has 33 attributes. The first attribute is a unique identification number, the second attribute is prognosis status (non-recurrence or recurrence) and the third attribute is recurrence time. The other 30 features were obtained by the process explained in the previous section. Two other features are the diameter of the removed tumor in centimeters and the number of axillary lymph nodes that were evaluated positively during surgery [21, 24]. The ten real valued and other features with statistical parameters of the WPBC dataset are shown in Table 4.
3.2 Classification Methods
Ten machine learning classification methods were used to diagnose breast cancer, recurrence or non-recurrence, benign or malignant using breast cancer datasets. Each algorithm is briefly described below.
3.2.1 AdaBoost
The ADB algorithm is a collective learning algorithm. In each iteration of this algorithm, a weak classifier is added. In each round t = 1, …, T. In each call, the weights are updated based on the importance of the samples. The weight of the incorrectly sorted samples is increased and the weight of the correctly sorted samples is reduced. As a result, the new classifier focuses on examples that are more difficult to learn [25].
3.2.2 Decision Tree
The DT algorithm is a hybrid algorithm and can perform classification operations on data [26]. A decision tree is made up of nodes. The director of a tree is a node called a “root” that has no input edges. All other nodes have exactly one input edge. A node with output edges is called an internal node or test node. Other nodes are called decision nodes or leaf. In a decision tree, each internal node divides the sample space into two or more spaces according to the values of the input attributes [27].
3.2.3 K nearest Neighbor
The KNN algorithm is a classification algorithm in which the class of a sample is categorized by a majority vote of its K neighbors. K is a positive value and is generally small. If k = 1, the sample is simply determined in its nearest neighbor class. K is basically considered an odd number to see an extra close neighbor and is prevented from equal votes [28].
3.2.4 Linear Discriminant Analysis
LDA involves finding the super plane and minimizing the variance between each class and maximizing the distance between the predicted mean classes. As a result, the linear composition can best find the attributes that separate two or more classes of objects. LDA is very close to the analysis of variance and regression analysis [29].
3.2.5 Logistic Regression
The LGR algorithm models the relationship between a variable that depends on the X classification and the Y attribute. In the LGR, there is a dependent variable and usually a set of independent variables which may be two categories, quantitative or a combination of them. The independent variable is small values and the qualitative dependent variable will have two values of zero or one [30].
3.2.6 Linear Regression
The LR algorithm measures the effect of an independent variable on a dependent variable and the correlation between them [31]. The common method for obtaining parameters is the least-squares method which the parameters are obtained by minimizing the sum of squares of error [32].
3.2.7 Naïve Bayes
The NB algorithm is a group of Naïve classifiers that use the Bayesian theorem as a strong assumption. In this method, by determining the probabilities of the results, uncertainty about the model is obtained. This classification is named after Thomas Bayes (1702–1761) who proposed this theorem [8, 33].
3.2.8 Artificial Neural Network
The ANN algorithm is a biological model. In this algorithm, a neural network can create the conditions for a computer to learn a problem that has not been seen before. The function of the ANN mimics the function of the human brain to some extent [34]. ANNs are made up of small units called neurons. The intensity of the connection between neurons is determined by synaptic weights. Each neuron can calculate the output value based on the weighted sum of the inputs [35].
3.2.9 Random Forest
The RF algorithm is a collective learning algorithm. For better decision-making, a set of DTs together produces a forest. Bagging is a technique used to generate training data in this algorithm. None of the selected data is deleted from the input dataset but is used to generate the next subset. In the bagging method, a random tree is created by creating N new data from the dataset. The final model is created by averaging or voting between the trees. In this method, the random tree is constructed in such a way that each time some variables are randomly selected and the best variable is selected from them [26]. Samples that are not selected in the training of trees in the bagging process are considered out of bag subsets and can be used to evaluate performance [36].
3.2.10 Support Vector Machine
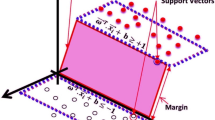
The SVM algorithm is one of the classification methods whose job is to select a line that has a maximum-margin hyperplane. Input space conversion is done implicitly using the Kernel function. If the data are not separated by a line, a hyperplane is created to categorize the data; So this hyperplane can have the highest margin compared to the samples in the classes [13].
3.3 Particle Swarm Optimization Algorithm
The PSO algorithm is one of the meta-heuristic algorithms. This algorithm was first developed in 1995 by Kennedy and Eberhart [11] and was initially used to simulate the mass flight of birds. After simplifying the initial algorithm, a kind of optimization operation was observed. Optimization is the process of improving performance in reaching the optimal point or points [37, 38].
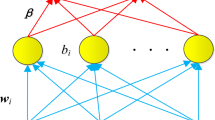
The PSO algorithm uses candidate solutions and a simple formula to solve the optimization problem and explores the search space of problem to obtain the optimal solution [11]. The usual aim of the PSO algorithm is to find the maximum or minimum of a function defined on a multidimensional vector space: for example find \(x^{*}\) such that \(f\left( {x^{*} } \right) < f\left( x \right)\) for all d-dimensional real vectors x. The objective function \(f:R^{d} \to R\) is called the fitness function. PSO is a swarm intelligence meta-heuristic inspired by the group behavior of animals, for example bird flocks or fish schools. Similarly to genetic algorithms (GAs), it is a population-based method, that is, it represents the state of the algorithm by a population, which is iteratively modified until a termination criterion is satisfied. In PSO algorithms, the population \(P = \left\{ {p_{1} , \ldots ,p_{n} } \right\}\) of the feasible solutions is often called a swarm. The feasible solutions \(p_{1} , \ldots ,p_{n}\) are called particles. The PSO algorithm views the set \(R^{d}\) of feasible solutions as a space where the particles move. For solving practical problems, the number of particles is usually chosen between 10 and 50.
3.3.1 Characteristics of Particle i at Iteration t
\(X_{i}^{\left( t \right)}\): The position (a d-dimensional vector) of ith particle at the iteration t.
\(Pbest_{i}^{\left( t \right)}\): The best solution that has been achieved so far by a particle (personal best).
\(Gbest^{\left( t \right)}\): The best solution that has been achieved so far by the entire swarm (global best).
\(V_{i}^{\left( t \right)}\): The speed of a particle.
At the beginning of the algorithm, the particle positions are randomly initialized, and the velocities are set to 0, or to small random values.
3.3.2 Parameters of the Algorithm
\(W^{(t)}\): Inertia weight; a damping factor, usually decreasing from around 0.9 to around 0.4 during the computation.
\(c_{1} , \, c_{2}\): The acceleration coefficients; usually between 0 and 2.
3.3.3 Update of the Speed and the Positions of the Particles
The velocity of particle i at the iteration t + 1 was obtained by updating the velocity of that particle at the previous iteration according to Eq. (1).
The symbols \(u_{1}\) and \(u_{2}\) represent random variables with the \(U\left( {0,1} \right)\) distribution. The first part of the velocity formula is called “inertia”, the second one “the cognitive (personal) component”, the third one is “the social (neighborhood) component”. Position of particle i at the iteration t + 1 was updated according to Eq. (2).
At each iteration, every particle should search around the minimum point it ever found as well as around the minimum point found by the entire swarm of particles. In other words in addition to position and velocity of the particles, the \(Pbest_{i}^{\left( t \right)}\) and \(Gbest^{\left( t \right)}\) was updated during the iterations.
3.3.4 Stopping Rule
The algorithm is terminated after a given number of iterations, or once the fitness values of the particles or the particles themselves are close enough in some sense. Figure 1 shows the flowchart of PSO algorithm.
3.3.5 Advantages of PSO Algorithm
In the PSO algorithm, a number of particles are randomly generated by the algorithm and they search for better solutions by moving in the domain of the problem. This is the similarity of PSO algorithm with genetic algorithm [39, 40]. There are very few algorithm parameters. The fitness function can be non-differentiable and only values of the fitness function are used. The method can be applied to optimization problems of large dimensions. The PSO algorithm is insensitive to scaling of design variables and it has a very efficient global search method.
4 Proposed PSO Feature Selection Algorithm
The aim of this research is to improve the accuracy of breast cancer diagnosis based on proposed PSO feature selection algorithm and machine learning classification methods. Figure 3 shows the flow chart of the proposed PSO feature selection algorithm. In the Data collection stage, three breast cancer datasets including Coimbra dataset, WDBC dataset, and WPBC dataset have been used. This process is done each time by one of the datasets. In the next step, after the data pre-processing steps are done, the PSO feature selection algorithm selects the most effective features in breast cancer diagnosis and produce a new dataset which has fewer features than the original dataset and whose dimensions have been reduced. Then, in the next step, the breast cancer diagnosis was done using new dataset which consists only of the features selected by the PSO feature selection algorithm, and using machine learning algorithms described in Sect. 3.2, which the data can be classified into two categories: sick or healthy. Also, using machine learning algorithms and original datasets without PSO feature selection algorithm, data classification into sick or healthy categories was done. At the end, the performance of machine learning methods with and without PSO feature selection algorithm are obtained and compared with each other. The detailed description of steps of the proposed PSO feature selection algorithm are as follows.
4.1 Data Pre-processing
In the data pre-processing phase, the dataset is transformed into understandable data. The tasks performed in the data preprocessing stage include:
-
1.
Detection and replacing the missing data
-
2.
Detection and replacing the outlier data
-
3.
Data normalization
-
4.
Data splitting.
The datasets were evaluated for missing value and outliers. Replacing missing and outlier values were done by replacing with the mean value of that variables.
Importing data without normalization reduce the accuracy and performance of machine learning models. For this reason, all input data were normalized between 0 and 1. Equation (3) was used to normalize the input data [41].
In this equation, min(x), max(x), and XN are the minimum, maximum and normalized data values of input data, respectively.
In the Data Splitting phase, the data were divided into two parts: Training and Test. Data dividing proportion were 80% for training and 20% for testing. To increase the generalizability of the model and prevent over fitting during model training, Cross-Validation (CV) technique was performed. In this method, the training data are divided into two parts. In an iterative process, in each iteration of cross validation method, one part of the data is used for the testing and the other parts for the training. The type of cross-validation performed was the K-fold method. In the K-fold CV method, the training dataset is randomly divided into k folds of the same size. At each iteration of the cross validation process, k − 1 of these folds can be used as the training dataset and one as the Validation dataset [42]. Figure 2 shows a tenfold cross-validation. In this study, a K-fold CV method with fourfold was implemented.
A tenfold cross validation [43]
Then, along with machine learning methods described in Sect. 4, once the datasets were evaluated without applying the PSO feature selection algorithm and again with applying PSO feature selection algorithm and using the more effective selected features by the PSO algorithm to diagnose breast cancer. After that, the results of these two approaches were compared (Fig. 3).
4.2 PSO Feature Selection Algorithm
The purpose of this research is to increase the accuracy of breast cancer diagnosis. The PSO feature selection objective function \(F(x)\) is shown in Eq. (4). According to this equation, the x vector is the inputs of the objective function which are the effective features selected by the PSO feature selection algorithm, and the output of the objective function is breast cancer diagnosis accuracy which is a summation of accuracy of training and validation dataset. Accuracy formula was described in the next section. Alpha and beta are coefficients of breast cancer diagnosis accuracy in training and validation datasets. The alpha and beta values was considered 0.7 and 0.3, respectively. In each iteration of the PSO feature selection algorithm, different features were selected by PSO algorithm and its fitness value which is breast cancer diagnosis accuracy was achieved by the objective function. Finally, after the termination of the iterations, the features selected by the PSO feature selection algorithm that have led the maximum accuracy are considered as the effective features in breast cancer diagnosis.
4.3 Performance Evaluation Parameters
For comparing the performance of different machine learning methods and different features selected by PSO feature selection algorithm, performance evaluation parameters included accuracy, sensitivity, specificity, F1_Score, positive predictive value (precision), negative predictive value, and area under the curve (AUC) was considered [44]. Figure 4 and Eqs. 5–10 show the confusion matrix for classification of two classes and the confusion matrix evaluation criteria, respectively. True positive (TP) is the number of cases correctly classified as patient. False positive (FP) is the number of cases incorrectly classified as patient. True negative (TN) is the number of cases correctly classified as healthy. False negative (FN) is the number of cases incorrectly classified as healthy. From the confusion matrix Accuracy, Sensitivity, Specificity and F1-score is evaluated using the following equations.
4.4 Sensitivity Analysis of Parameters of PSO Algorithm and Machine Learning Methods
In this research, the default values available in the Sklearn library of the Python programming language have been used for the parameters of machine learning algorithms. In order to choose the appropriate values of PSO algorithm parameters, a sensitivity analysis based on accuracy, sensitivity and specificity was performed using one of the machine learning methods. Tables 5, 6, 7 and 8 describe the sensitivity of the PSO algorithm to the variations in its parameters.
According to the results obtained in the above tables, the values of the parameters for PSO feature selection algorithm have been considered as described in Table 9.
All the process of breast cancer diagnosis and PSO feature selection algorithm along with machine learning methods were done using python 3 programming language on a PC with Intel (R) Core ™ i5-540 2.53GHz CPU and 8.00 GB of RAM.
5 Results
This section presents the breast cancer diagnosis results obtained from the classification evaluation with and without PSO feature selection for the three datasets Coimbra, WDBC and WPBC.
5.1 Classification Evaluation for Coimbra Dataset
In without PSO feature selection, ten machine learning models were constructed with all features in the Coimbra dataset. In PSO feature selection, ten machine learning models were constructed with selected features by PSO algorithm. The results after comparing these two phases are shown in Figs. 5 and 6, respectively. It is clear from Figs. 5 and 6 that PSO feature selection can improve diagnosis of breast cancer disease with increase accuracy, sensitivity and specificity.
In Fig. 7, the AUC evaluation metric is compared in the PSO feature selection and without PSO feature selection. According to the results, it is clear that the PSO feature selection could improve the AUC value.
Breast cancer diagnosis results without PSO feature selection and using PSO feature selection are shown in Tables 10 and 11, respectively. Using proposed PSO feature selection algorithm the accuracy of ADB, DT and LDA models improved by 3% and the accuracy of RF, LR, and LGR models improved by 2%. The accuracy of KNN, NB, NN and SVM models improved by 19, 8, 6 and 4%, respectively, using PSO feature selection algorithm compared to without PSO feature selection.
According to Fig. 8, the experimental results show that the prediction model based on SVM classifier and PSO feature selection algorithm has a higher accuracy percentage to predict the factors influencing cancer breast prediction compared to other algorithms.
Figure 9 also shows the number of features selected with and without PSO feature selection for each ML models in Coimbra dataset. This dataset consists of nine features. Using the PSO feature selection algorithm, the number of features of ADB and SVM models was reduced to four features, and the number of features of DT, KNN, LDA, LR and NN models was reduced to six features. The number of features of LGR, NB, RF models was reduced to five features.
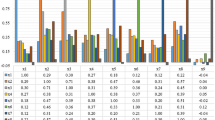
In addition, classification evaluation metrics are given for ten ML models in Fig. 10.
5.2 Classification Evaluation for WDBC Dataset
Similar to the previous dataset, constructed ten machine learning models without PSO feature selection and with PSO feature selection in WDBC dataset. Comparing results of these two phase are shown in Figs. 11 and 12, respectively.
Figure 13 shows the comparison of AUC in with and without PSO feature selection. With this comparison, it is clear that the AUC value has improved in the PSO feature selection.
The diagnostic results of benign or malignant breast cancer for all features without PSO feature selection and using PSO feature selection are given in Tables 12 and 13, respectively. Using PSO feature selection algorithm the accuracy of ADB, DT, KNN, LR, NN, RF models improved by 1% and the accuracy of NB and SVM models improved by 3% and 2%, respectively. The accuracy for LDA and LGR models were the same with and without using PSO feature selection algorithm but for LDA model the sensitivity has improved by 1% with using the PSO feature selection. The classification results for LGR model with and without PSO feature selection were the same.
According to Fig. 14, the diagnostic result of the ADB classifier with the PSO feature selection algorithm has a higher accuracy percentage compared to other ML models.
Figure 15 shows the number of features selected with and without PSO feature selection for each ML models in WDBC dataset. This dataset consists of 30 features. By using the PSO feature selection algorithm, the number of features of ADB and LR models was reduced to 21 features and the number of features of DT and LDA models was reduced to 14 features. The number of features of KNN and SVM models was reduced to 16 features and the number of features of LGR, NB, NN, RF models was reduced to 9, 15, 17 and 19 features, respectively.
In addition, classification evaluation metrics are given for ten ML models in Fig. 16.
5.3 Classification Evaluation for WPBC Dataset
Similar to the previous two datasets, constructed ten machine learning models without PSO feature selection and with PSO feature selection. Comparing results of these two phase are shown in Figs. 17 and 18, respectively.
In Fig. 19, the AUC of with and without PSO feature selection is compared. The AUC has been improved when PSO feature selection is used.
The breast cancer recurrence or non-recurrence diagnosis results for all features without PSO feature selection and using PSO feature selection is given in Tables 14 and 15, respectively.
Using proposed PSO feature selection algorithm the accuracy of DT, LGR, NN, RF and SVM models improved by 2%. Also, the accuracy of ADB and NB models improved by 3%, the accuracy of LDA and LR models improved by 1% and the accuracy of KNN model improved by 4%.
According to Fig. 20, the recurrence diagnosis result of SVM classifier with PSO feature selection algorithm has a higher accuracy percentage compared to other ML models.
Figure 21 shows the number of features selected with and without PSO feature selection for each ML model in WPBC dataset. This dataset consists of 33 features. Using the PSO feature selection algorithm, the number of features of ADB, DT, NB and RF models was reduced to 20 features and the number of features of LGR and NN models was reduced to 18 features.The number of features of KNN, LDA, LR and SVM models was reduced to 19, 21, 23, and 17 features, respectively.
Classification evaluation metrics are given for ten machine learning models in Fig. 22.
6 Discussion
In this study, performance evaluation metrics including accuracy, sensitivity, specificity, precision, and AUC have been used.
According to the results, when the PSO feature selection is used, all classifiers perform better in terms of accuracy level than when PSO feature selection. In some classifier, the accuracy obtained is close or equal with or without PSO feature selection. But in the PSO feature selection, the results were obtained with a smaller number of features compared to all features in the datasets (Tables 16, 17).
Also, classifiers have a higher level of precision with PSO feature selection compared to without PSO feature selection (Figs. 23, 24 and 25).
In the PSO feature selection, better or equal results are obtained than selecting all features Figure 26 compares the accuracy in the PSO feature selection between the three datasets. According to this figure, the best result of the accuracy level for diagnosis of breast cancer in Coimbra dataset, obtained by the SVM classifier with an accuracy of 91%. After that ADB had the best result with an accuracy of 88%. In the WDBC dataset, the best result obtained by RF classifier with an accuracy of 100% and then ADB classifier with an accuracy of 99%. In the WPBC dataset, the best result obtained by the SVM classifier with an accuracy of 96% and then RF classifier with an accuracy of 95%. Therefore, feature selection reduces the size of the data and improves the results.
6.1 Comparing the Results of Classifiers in Each Dataset
In this section, a comparative analysis of all evaluation metrics and their measurement for each classifier in each dataset with and without PSO feature selection was made.
When using the PSO feature selection, the accuracy result of all classifiers in three datasets were improved. In some classifiers the accuracy of PSO feature selection may be equal to without PSO feature selection, but in this cases the accuracy of PSO feature selection is obtained with significantly less number of features compared to all features and this is a great advantage of PSO feature selection.
In Coimbra dataset with 9 features, PSO feature selection algorithm improved the accuracy of breast cancer diagnosis by 2, 3, 10, 9, 8, 2, 6, 2, 4% in ADB, DT, KNN, LDA, LGR, LR, NB, NN, RF, SVM classifiers by selecting 4, 6, 6, 6, 5, 6, 5, 6, 5, 4 number of features, respectively (Fig. 27).
In WDBC dataset with 30 features, PSO feature selection algorithm improved the accuracy of breast cancer malignant or benign diagnosis by 1, 1, 1, 0, 0, 1, 1, 1, 2% in ADB, DT, KNN, LDA, LGR, LR, NB, NN, RF, SVM classifiers by selecting 21, 14, 16, 14, 9, 21, 15, 17, 19, 16 number of features, respectively (Fig. 28).
In WPBC dataset with 33 features, PSO feature selection algorithm improved the accuracy of breast cancer malignant or benign diagnosis by 3, 2, 4, 1, 2, 1, 3, 2, 3, 2% in ADB, DT, KNN, LDA, LGR, LR, NB, NN, RF, SVM classifiers by selecting 20, 20, 19, 21, 18, 23, 20, 18, 20, 17 number of features, respectively (Fig. 29).
Other evaluation metrics revealed that using PSO feature selection algorithm can improve their results too.
6.2 Comparison the Results with the Related Works
Table 18 shows the results of related works along with the parameters and models used in them. This research has been able to achieve a higher level of accuracy than these similar works.
7 Conclusion and Future Work
Breast cancer is one of the leading cause of death of women in the world. There are a large number of features involved in correctly diagnosis of this disease. For this reason, reducing the number of these features and finding the effective features related to breast cancer diagnosis can help the patients and speed up the process of diagnosing. This article focused on improving the accuracy of ten machine learning classification methods by integrating Particle swarm optimization (PSO) feature selection algorithm in to them. Using PSO feature selection (FS) algorithm, machine learning classification methods are improved and the number of features is reduced. The proposed method evaluated with three common breast cancer dataset named Coimbra, WDBC and WPBC datasets. The study results showed that in the Coimbra dataset with 9 features, PSO feature selection algorithm improved the accuracy of breast cancer diagnosis by 2, 3, 10, 9, 8, 2, 6, 2, 4% in AdaBoost (ADB), Decision Tree (DT), k Nearest Neighbors (KNN), Linear Discriminant Analysis (LDA), Logistic Regression (LR), Linear Discriminant Analysis (LDA), Naïve Bayes (NB), Artificial Neural Networks (NN), Random Forest (RF), Support Vector Machine (SVM) classifiers by selecting 4, 6, 6, 6, 5, 6, 5, 6, 5, 4 number of features, respectively. In this dataset, the best accuracy of 91% was obtained by the SVM classifier integrated with PSO feature selection algorithm. In WDBC dataset with 30 features, PSO feature selection algorithm improved the accuracy of breast cancer malignant or benign diagnosis by 1, 1, 1, 0, 0, 1, 1, 1, 2% in ADB, DT, KNN, LDA, LGR, LR, NB, NN, RF, SVM classifiers by selecting 21, 14, 16, 14, 9, 21, 15, 17, 19, 16 number of features, respectively. In this dataset, the best accuracy of 100% was obtained by the RF classifier integrated with PSO feature selection algorithm. In WPBC dataset with 33 features, PSO feature selection algorithm improved the accuracy of breast cancer malignant or benign diagnosis by 3, 2, 4, 1, 2, 1, 3, 2, 3, 2% in ADB, DT, KNN, LDA, LGR, LR, NB, NN, RF, SVM classifiers by selecting 20, 20, 19, 21, 18, 23, 20, 18, 20, 17 number of features, respectively. In this dataset, the best accuracy of 96% was obtained by the SVM classifier integrated with PSO feature selection algorithm. The results of this research indicates that the PSO feature selection algorithm could identify most effective features in breast cancer diagnosis. Therefore, the disease could be diagnose with higher accuracy in a shorter time and save patient’s lives. It is recommended to use other machine learning methods such as other ensemble techniques and other meta-heuristic feature selection algorithms and compare their results with the PSO feature selection algorithm.
Availability of Data and Material
Three datasets including Coimbra dataset, WDBC dataset, and WPBC dataset have been used. These datasets are published on the UCI Machine Learning Repository website with public access.
Abbreviations
- ADB:
-
AdaBoost
- ANN:
-
Artificial neural networks
- DT:
-
Decision tree
- ELM:
-
Extreme learning machine
- FNA:
-
Fine needle aspirate
- FS:
-
Feature selection
- KNN:
-
k nearest neighbors
- RF:
-
Random forest
- SVM:
-
Support vector machine
- LDA:
-
Linear discriminant analysis
- LGR:
-
Logistic regression
- LR:
-
Linear regression
- NB:
-
Naïve Bayes
- UCI:
-
University of California, Irvine
- PSO:
-
The particle swarm optimization
- SI:
-
Swarm intelligence
- WDBC:
-
Wisconsin Diagnostic Breast Cancer dataset
- WPBC:
-
Wisconsin Prognostic Breast Cancer dataset
References
Salem, A., Sherif, A.S., Hussein, M.K.: Efficient framework for detecting COVID-19 and pneumonia from chest X-ray using deep convolutional network. Egypt. Inform. J. 23(2), 247–257 (2022). https://doi.org/10.1016/j.eij.2022.01.002
Salehiniya, H., Ghoncheh, M., Pournamdar, Z.: Incidence and mortality and epidemiology of breast cancer in the world. Asian Pac. J. Cancer Prev. 17, 43–46 (2016)
Najjar, H., Easson, A.: Age at diagnosis of breast cancer in Arab nations. Int. J. Surg. 8(6), 448–452 (2010). https://doi.org/10.1016/j.ijsu.2010.05.012
El Saghir, N.S., et al.: Trends in epidemiology and management of breast cancer in developing Arab countries: a literature and registry analysis. Int. J. Surg. 5(4), 225–233 (2007). https://doi.org/10.1016/j.ijsu.2006.06.015
Musallam, A.S., Sherif, A.S.: A new convolutional neural network architecture for automatic detection of brain tumors in magnetic resonance imaging images. IEEE Access 10, 2775–2782 (2022). https://doi.org/10.1109/ACCESS.2022.3140289
Chakradeo, K., Vyawahare, S., Pawar, P.: Breast cancer recurrence prediction using machine learning. In: 2019 IEEE Conf. Inf. Commun. Technol. CICT 2019, pp. 1–7 (2019). https://doi.org/10.1109/CICT48419.2019.9066248
Vijayalakshmi, et al.: Multi-modal prediction of breast cancer using particle swarm optimization with non-dominating sorting. Int. J. Distrib. Sens. Netw. (2020). https://doi.org/10.1177/1550147720971505
Sakri, S.B., Abdul Rashid, N.B., Muhammad Zain, Z.: particle swarm optimization feature selection for breast cancer recurrence prediction. IEEE Access 6, 29637–29647 (2018). https://doi.org/10.1109/ACCESS.2018.2843443
Nurhayati, F., Agustian, Lubis, M.D.I.: Particle swarm optimization feature selection for breast cancer prediction. In: 2020 8th Int. Conf. Cyber IT Serv. Manag. CITSM 2020, pp. 5–10 (2020). https://doi.org/10.1109/CITSM50537.2020.9268865
Akben, S.B.: Determination of the blood, hormone and obesity value ranges that indicate the breast cancer, using data mining based expert system. IRBM 1, 1–6 (2019). https://doi.org/10.1016/j.irbm.2019.05.007
Kennedy, J.: Particle swarm: social adaptation of knowledge. In: Proc. IEEE Conf. Evol. Comput. ICEC, pp. 303–308 (1997). https://doi.org/10.1109/icec.1997.592326
Mafarja M., Sabar, N.R.: Rank based binary particle swarm optimisation for feature selection in classification. In: ACM Int. Conf. Proceeding Ser., no. June (2018). https://doi.org/10.1145/3231053.3231072
Silva, S., Anunciação, O., Lotz, M.: A comparison of machine learning methods for the prediction of breast cancer. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 6623 LNCS, pp. 159–170 (2011). https://doi.org/10.1007/978-3-642-20389-3_17
Silva Araújo, V., Guimarães, A., de Campos Souza, P., Silva Rezende, T., Souza Araújo, V.: Using resistin, glucose, age and bmi and pruning fuzzy neural network for the construction of expert systems in the prediction of breast cancer. Mach. Learn. Knowl. Extr. 1(1), 466–482 (2019). https://doi.org/10.3390/make1010028
Malla, Y.A.: A machine learning approach for early prediction of breast cancer. Int. J. Eng. Comput. Sci. (2017). https://doi.org/10.18535/ijecs/v6i5.31
Patrício, M., et al.: Using resistin, glucose, age and BMI to predict the presence of breast cancer. BMC Cancer (2018). https://doi.org/10.1186/s12885-017-3877-1
Paquin, F., Rivnay, J., Salleo, A., Stingelin, N., Silva, C.: Breast cancer diagnosis by different machine learning methods using blood analysis data. J. Mater. Chem. C 3(January 2019), 10715–10722 (2018). https://doi.org/10.1039/b000000x
Wang, H., Zheng, B., Yoon, S.W., Ko, H.S.: A support vector machine-based ensemble algorithm for breast cancer diagnosis. Eur. J. Oper. Res. 267(2), 687–699 (2018). https://doi.org/10.1016/j.ejor.2017.12.001
2013 UCI: UC Irvine machine learning repository (2013)
UCI machine learning repository, breast cancer Coimbra data set (2018). https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Coimbra.
Maglogiannis, I., Zafiropoulos, E., Anagnostopoulos, I.: An intelligent system for automated breast cancer diagnosis and prognosis using SVM based classifiers. Appl. Intell. 30(1), 24–36 (2007). https://doi.org/10.1007/s10489-007-0073-z
UCI machine learning repository, Breast Cancer Wisconsin (Diagnostic) data set (1995). https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+(Diagnostic).
Wolberg, W.H., Mangasarian, O.L.: Multisurface method of pattern separation for medical diagnosis applied to breast cytology. Proc. Natl. Acad. Sci. U.S.A. 87(23), 9193–9196 (1990). https://doi.org/10.1073/pnas.87.23.9193
Breast Cancer Wisconsin (Prognostic) Data Set (1995). https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+(Prognostic).
Freund, Y., Schapire, R.E.: A decision-theoretic generalization of on-line learning and an application to boosting. Lect. Not. Comput. Sci. (including Subser. Lect. Not. Artif. Intell. Lect. Not. Bioinform.) 904, 23–37 (1995). https://doi.org/10.1007/3-540-59119-2_166
Schapire, R.E.: Random Forests. 2001 Kluwer Acad. Publ. Manuf. Netherlands., vol. 2012, no. 4, pp. 1–7 (2001). http://ci.nii.ac.jp/naid/110009490689/
Galitskaya, E.G., Galitskkiy, E.B.: Classification trees. Sotsiologicheskie Issled. 3, 84–88 (2013). https://doi.org/10.4018/978-1-60960-557-5.ch006
Kramer, O.: Dimensionality reduction with unsupervised nearest neighbors. Intell. Syst. Ref. Libr. 51, 13–23 (2013). https://doi.org/10.1007/978-3-642-38652-7
Xanthopoulos, P., Pardalos, P.M., Trafalis, T.B.: Robust Data Mining, pp. 27–33. Springer, New York (2013). https://doi.org/10.1007/978-1-4419-9878-1
Ayyadevar, V.K.: Pro Machine Learning Algorithms, pp. 49–69. Apress, Berkeley (2018). https://doi.org/10.1007/978-1-4842-3564-5
Rencher, A.C.: A review of “methods of multivariate analysis, second edition.” IIE Trans. 37(11), 1083–1085 (2005)
Zhu, Y., Zhu, C., Li, X.: Improved principal component analysis and linear regression classification for face recognition. Signal Process. 145, 175–182 (2018). https://doi.org/10.1016/j.sigpro.2017.11.018
Webb, G.I.: Naïve Bayes. Encycl. Mach. Learn., vol. 국내석사학위논문 (2012). http://www.riss.kr/link?id=T12754991.
Cartwright, H.: Artificial Neural Networks, 2nd edn. Springer, New York (2014)
Dreyfus, G.: Neural Networks. Springer, Berlin (2005)
Peters, J., et al.: Random forests as a tool for ecohydrological distribution modelling. Ecol. Modell. 207(2–4), 304–318 (2007). https://doi.org/10.1016/j.ecolmodel.2007.05.011
Abu, O., Abo-hammour, Z.: Numerical solution of systems of second-order boundary value problems using continuous genetic algorithm. Inf. Sci. (NY) (2014). https://doi.org/10.1016/j.ins.2014.03.128
Abo-hammour, Z., Arqub, O.A., Momani, S., Shawagfeh, N.: Optimization solution of Troesch’s and Bratu’s problems of ordinary type using novel continuous genetic algorithm. Discrete Dyn. Nat. Soc. 2014(1), 401696 (2014)
Abo-hammour, Z., Alsmadi, O., Momani, S., Arqub, O.A.: A genetic algorithm approach for prediction of linear dynamical systems. Math. Probl. Eng. 2013, 1–12 (2013)
Arqub, O.A., Abo-hammour, Z., Momani, S., Shawagfeh, N.: Solving singular two-point boundary value problems using continuous genetic algorithm. Abstr. Appl. Anal. (2012). https://doi.org/10.1155/2012/205391
Saleh, H.: Machine Learning Fundamentals: Use Python and Scikit-Learn to Get Up and Running with the Hot-Test Developments in Machine Learning, pp. 1–37. Packt Publishing, Birmingham (2018)
Abdar, M., et al.: A new nested ensemble technique for automated diagnosis of breast cancer. Pattern Recognit. Lett. 132, 123–131 (2020). https://doi.org/10.1016/j.patrec.2018.11.004
KarlRosaen: K-fold cross-validation (2016). http://karlrosaen.com/ml/learning-log/2016-06-20/.
Zhou, Z.H.: Cost-sensitive learning. In: Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics), vol. 6820 LNAI, pp. 17–18 (2011) https://doi.org/10.1007/978-3-642-22589-5_2.
Li, Y.: Performance evaluation of machine learning methods for breast cancer prediction. Appl. Comput. Math. 7(4), 212 (2018). https://doi.org/10.11648/j.acm.20180704.15
Funding
No funds, grants, or other support was received.
Author information
Authors and Affiliations
Contributions
Reihane Kazerani carried out this research.
Corresponding author
Ethics declarations
Conflict of interest
The author declares that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Consent for publication
The author agrees the article to be published.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kazerani, R. Improving Breast Cancer Diagnosis Accuracy by Particle Swarm Optimization Feature Selection. Int J Comput Intell Syst 17, 44 (2024). https://doi.org/10.1007/s44196-024-00428-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44196-024-00428-5