Abstract
A deep convolution neural network image segmentation model based on a cost-effective active learning mechanism is proposed and named PolySeg Plus. It is intended to address polyp segmentation with a lack of labeled data and a high false-positive rate of polyp discovery. In addition to applying active learning, which assisted in labeling more image samples, a comprehensive polyp dataset formed of five benchmark datasets was generated to increase the number of images. To enhance the captured image features, the locally shared feature method is used, which utilizes the power of employing neighboring features together with one another to improve the quality of image features and overcome the drawbacks of the Conditional Random Features method. Medical image segmentation was performed using ResUNet++, ResUNet, UNet++, and UNet models. Gaussian noise was removed from the images using a gaussian filter, and the images were then augmented before being fed into the models. In addition to optimizing model performance through hyperparameter tuning, grid search is used to select the optimum parameters to maximize model performance. The results demonstrated a significant improvement and applicability of the proposed method in polyp segmentation when compared to state-of-the-art methods on the datasets CVC-ClinicDB, CVC-ColonDB, ETIS Larib Polyp DB, KVASIR-SEG, and Kvasir-Sessile, with Dice coefficients of 0.9558, 0.8947, 0.7547, 0.9476, and 0.6023, respectively. Not only did the suggested method improve the dice coefficients on the individual datasets, but it also produced better results on the comprehensive dataset, which will contribute to the development of computer-aided diagnosis systems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Colorectal cancer is recognized as a dangerous disease causing deaths worldwide, with nearly two million new cases and 1 million cancer deaths in the last 2 years [1]. Like any type of cancer, healthy human body cells can turn into harmful cells in the form lesions [2]. Colorectal cancer commonly arises from polyps of the colon or rectal epithelium, which are non-cancerous neoplasms. Some polyps can develop into precancerous lesions, which can lead to colorectal cancer. Detecting and removing adenomas early (early screening) will reduce the severity of colorectal cancer. In the USA, colorectal cancer represents the third most common reason causing cancer for men and women and the second reason causing deaths for both genders [3].
Colorectal cancer, which is also called bowel cancer, has several risk factors that have been approved by the American Cancer Society. The most important risk factors are lifestyle-related and changeable, such as being overweight or obese, not being physically active, certain types of diet, and alcohol consumption. On the contrary, there are other factors that cannot be changed over time, such as one’s age [4], history of a certain person or one of his family members with polyps, cancer, or inflammatory bowel disease [5], as well as an inherited syndrome.
The Adenomas Detection Rate (ADR) measures the frequency with which a practitioner detects precancerous adenomas. A 1% rate is considered a good adenoma detection rate, which is accompanied by a 3% reduction in the risk of having colorectal cancer [6, 7]. This rate is thought to be influenced by two aspects: blind spots and human error. The first aspect could be addressed using a broad scope, while the second aspect is challenging, and researchers are very interested in artificial intelligence to reduce human error.
A medical endoscopy decision support system follows a standard procedure. The first step is often to prepare the tissue region to be studied. Preprocessing may be required after an image has been acquired to improve the quality of degraded photos. Based on the application’s goal [8], the appropriate features must then be located and extracted to detect polyps or cancer. Some methods, like classification, are intended for Content-Based Image Retrieval (CBIR) [9] or Content-Based Video Retrieval (CBVR). The primary distinction between automated decision support systems and CBIR/CBVR systems is that the output of a decision support system based on automation [10] can be a suggestion for the last diagnosis phase or more information for a diagnosis.
Medical image segmentation is the process of extracting Regions of Interest (ROIs) from 3D image data such as Magnetic Resonance Imaging (MRI) or Computed Tomography (CT) scans [11]. The primary goal of the segmentation task is to highlight areas of the anatomy needed for a specific study. Segmentation of images consumes much time, but recent advances in Artificial Intelligence (AI) tools are trying to make repetitive tasks faster and more efficient.
The problem is to detect and remove precancerous adenomas in patients with colorectal cancer, which significantly reduces the severity of the disease. Factors such as lifestyle risks and genetic syndromes contribute to the development of the disease. Therefore, more efficient and accurate detection methods are needed to reduce human error and eliminate screening blind spots. A medical endoscopy decision support system using AI tools and image segmentation techniques may improve adenoma detection rates and reduce the impact of colorectal cancer.
The novelty and main work of this paper are as follows:
-
1.
Reducing the high false-positive rates of polyp discovery in SOTA algorithms.
-
2.
Enhancing and improving the image quality in the preprocessing phase using Gaussian filters.
-
3.
Contributing to the shortage of labeled data (normal images without polyps) problem by applying a cost-effective active learning technique.
-
4.
Creating a comprehensive polyp dataset by combining six different datasets.
-
5.
Applying Locally Shared Featured technique and integrating it with deep learning models to improve their performance and reduce computational time.
-
6.
Hyperparameter tuning using grid search to enhance the performance of the models.
The proposed study aims to develop an automated system to help gastroenterologists segment polyps of various sizes and decide whether to remove or leave the polyp after examination. A Gaussian filter is used in the preprocessing stage to improve the image quality. We combine six different data sets to create a comprehensive polyp data set and use active learning techniques to address the lack of normal labeled data (images without polyps). Grid search hyperparameter tuning is performed to select the best parameters and optimize the model. The ultimate goal is to improve the accuracy and efficiency of polyp segmentation, giving gastroenterologists better information to make informed polypectomy decisions.
The rest of the paper is organized as follows: Sect. 2, introduces a brief introduction to medical image segmentation techniques. Section 3 describes the related work done in polyp segmentation. Section 4 illustrates the datasets and various methods, while Sect. 5 shows the different variations of the proposed model architecture used in this study. Section 6 presents the results and experiments of the proposed model and state-of-the-art (SOTA) models, in addition to ablation studies. Section 7 discusses the experiment results in detail compared to previous work. Section 8 discusses the hypothesis and the limitations of the proposed model. Finally, Sect. 9 is the study’s conclusion.
2 Background
The purpose of this section is to set the context and provide the foundation for understanding medical image segmentation, computer-aided diagnosis, and their relation to the deep learning field, that helps demonstrate the important terms in the existing knowledge.
2.1 Medical Image Segmentation
One of the key benefits of medical image segmentation is that it facilitates much more specific anatomical analysis of data by separating only the areas that are required [12]. Segmentation works with CT, MRI, as well as other types of scans by producing a mask from the background image data. Based on the task, users are able to work on their scans in two or three dimensions colorectal polyp segmentation is indeed a difficult task caused by variations in polyp form and color intensity in colonoscopic frames [13]. Polyp segmentation was divided into three main methods by the researchers. The first method is image processing-based segmentation, which does not employ any learning methods. The second method involves extracting features first and then segmenting them using classifiers, as shown in Fig. 1 where on the left side is the raw image, which is considered an input to the model, while on the right side is the output, which is the segmented image or ground truth (mask). In the third method, approaches that perform segmentation using convolutional neural networks are grouped together.
2.2 Computer-Aided Diagnosis (CAD)
Computer-aided detection is a computer-based framework that assists medical physicians in making quick decisions in the field of medical imaging [14]. Medical imaging is concerned with information existing in images that medical practitioners, such as gastroenterologists, must assess and analyze in a short span of time, such as discovering polyps that will aid in the decision of whether to leave or resect these polyps. Image processing evaluation is an important task in the medical sector because imaging is a basic method for identifying any disease in its early phases, but image acquisition should also not endanger the human body, such as during endoscopic operations, X-ray, and MRI scans [15], and so on. Images taken with great intensity of energy provide superior quality but endanger the body; thus, images are captured with much less energy and in turn, will have poor quality and low contrast, which will be a valuable area to investigate by the researchers.
2.3 Deep Learning
Deep learning and machine learning are the foundations of any CAD or medical decision support system. Deep learning is based on combining low-level features, placing higher-level abstract feature characteristics, and classifying intangible objects. Deep learning methodology is derived by researchers from different studies and experiments on artificial neural networks. The most used deep learning models for processing and analyzing images are convolutional neural networks (CNNs) or deep convolutional neural networks (DCNNs), in addition to the recurrent neural network (RNN), model which is widely used in CNNs, with different network frameworks, such as long short-term memory (LSTM) networks, a form of recurrent neural networks that is good at learning order reliance in predicting sequence [16].
3 Literature Review
The purpose of this section is to highlight the progress made, identify current problems, and establish the way for creative approaches to the accurate segmentation and localization of polyps by conducting a comprehensive review of earlier works.
In 2020, Mandal et al. [17] developed a reliable and effective method for segmenting polyp regions. In their research, fuzzy clustering was used to split polyp areas from healthy areas in colonoscopic image frames by producing a distinctive threshold level from the hue, saturation, and lightness color space’s V channel. There are several types of fuzzy clustering that investigate cluster information, including hard and soft clustering. Hard clustering divides the data into distinct groups or distinct clusters, with each data object precisely assigned to one of the groups. Soft clustering, on the other hand, assigns each data object to one or perhaps more clusters, with membership levels assigned during the process. Their model achieved an accuracy of 98.80% compared to three other studies proposed by Hwang et al., Alexandre LA et al., and Kodogiannis et al., where their accuracy are 77.77%, 94.87% and 97.14%, respectively.
In 2021, Debesh Jha et al. [18] did thorough research into segmenting colorectal polyps. They used many different models such as ResUNet++, ResUNet, and UNet as major models. These models were tested on six datasets, which are CVC-ClinicDB, CVC-ColonDB, ETIS Larib Polyp DB, Kvasir-SEG, ASU-Mayo Clinic Colonoscopy Video Database and CVC-VideoClinicDB, with a total of 33,119 images. Data augmentation was applied to increase the number of polyps, and they also reduced the complexity by modifying the size of the images to 256 \(\times \) 256. They improved the results of the experimental model by implementing augmentation at the test time and Conditional Random Field (CRF) as a post-processing technique. After testing the proposed model on different datasets, they concluded that ResUNet++ is better at segmenting all different types of polyps (large, small, and regular polyps), especially smaller and sessile polyps. In addition, using ResUNet++ combined with the CRF improved precision and recall.
In 2021, Banik et al. [19] developed a polyp segmentation network called Polyp-Net that is based on fusion. They enhanced the CNN with a network called a Binary Tree Wavelet. The dataset used in this study is from a polyp segmentation challenge called Endoscopic Vision held in Singapore. For training, they used 300 frames, while for testing, they used 612 frames. In the preprocessing phase, they focused on noise in the frames such as blood vessels and endoluminal folds by applying the Mumford-Shah-Euler in-painting method. Since the resulting segmented image was not promising in terms of an accurate region of interest, they used Local Gradient Weighting as a type of Level-Set Method (LSM) to overcome this problem. Their proposed model outperformed CNN and achieved a precision and recall of 0.836 and 0.811, respectively, compared to UNet and ResNet-50.
In 2022, Qiu et al. [20] designed the Boundary Distribution Guided Network (BDG-Net) to segment polyps accurately. The research focused on enhancing segmentation by integrating many scale features since polyps have various sizes and undefined boundaries. The suggested model consists of two units. The first unit is for generating boundary distribution, which is used to assemble high-level features and generate a map of this boundary. The second unit is the Boundary Distribution Guided Decoder (BDGD), which enhances polyp segmentation using the previously generated BDM and integrates that with many scale features. The training set contained a total of 1450 images from CVC-ClinicDB and Kvasir, while they used three different datasets for testing, which are CVC300, ETIS, and CVC-ColonDB. They contrasted their proposal with the state-of-the-art algorithms such as SFA, PraNet, UNet, UNet++, ResUNet-mod, and ResUNet++. The proposed method achieved a mean dice of 0.915, which outperformed the previously mentioned algorithms.
In 2022, Mohapatra et al. [21] proposed a segmentation architecture called U-PolySeg that concatenates features using dilated convolution. Due to their different sizes, the images were resized to 416 \(\times \) 416 pixels during the processing step. A comprehensible transport module was applied to remove specular reflections in the image, and the contrast of the images was enhanced using contrast limited adaptive histogram equalization. The architecture of UNet model was modified to add more advanced blocks. Many experiments were done to select the best parameters of the proposed model to ensure its effectiveness. The dataset used was the Kvasir-SEG dataset, which has 1000 images and masks. Finally, they compared their proposed model to ColonSegNet. The proposed model achieved 0.9677, 0.9686, 0.8791, 0.9557, and 0.9229 in terms of global accuracy, dice coefficient, intersection over union, recall, and precision, respectively.
In 2022, Gautam et al. [22] constructed an encoder and decoder structure and focused on multi-scale features by applying squeeze and excitation modules. They modified the skip connection by using Fusion Attention Blocks to minimize the semantic gap between both encoder and decoder (FAB). To enrich and extract more features, a Multi-Scale Information (MSI) block is applied, which will help in the representation of relevant features. The Kvasir-SEG dataset was used for training and testing, and due to the lack of labeled data, data were augmented using a special library called albumentations. The proposed model succeeded in segmenting different sizes of polyps and achieved a dice score of 85.15% compared to the other four models.
In 2022, Tran et al. [23] proposed a model that is an output of a modification in the residual recurrent UNet architecture. The new model was implemented to minimize the size of the model and the change convolutional filters in a more flexible way; they named the new model the Modified Residual Recurrent UNet model (MRR-UNet). Two variations of (MRR-UNet) were implemented: mRR1-UNet and mRR2-UNet, the first version consisted of 16 filters, while the second version consisted of 32 filters. The datasets included were three datasets: CVC-ColonDB, ETIS-LaribPolypDB, and CVC-ClinicDB. For training purposes, all images were resized to \(224 \times 224\). Data augmentation was used because of the limitations of the image numbers. Augmentation methods were used such as shearing, rotating, and flipping images. Their model achieved an average dice of 93.54%.
To the best of our knowledge, the models mentioned in previous research used various techniques to segment polyps, but these studies ignored many aspects, such as improving image quality and applying different processing techniques that will improve model results, as well as an important performance measure such as recall, in addition to the limited amount of medical data.
In summary, there have been several research studies in recent years that have attempted to develop reliable and effective methods for segmenting polyp regions in colonoscopic images. These studies have used various techniques such as fuzzy clustering, Binary Tree Wavelet, Local Gradient Weighting, Boundary Distribution Guided Network. These methods have been tested on different datasets, with varying levels of success. Some have achieved higher accuracy, precision, and recall compared to other studies and state-of-the-art algorithms. However, the best method for polyp segmentation may depend on the specific dataset and application. Table 1 summarizes the literature studies as follows: dataset, preprocessing, methodology, evaluation tools, advantages, and disadvantages
In this study, we propose an enhancement to overcome existing research gaps by designing a versatile and robust model that is independent of a specific dataset. Additionally, previous studies did not include the use of normal images to address the shortage of normal labeled training data. To address this, we implement Cost-Effective Active Learning to increase the number of normal images. To further improve the results, we enhance images using Gaussian filters. In contrast to a previous study that used CRF as a post-processing technique, we utilize Locally Shared Features to improve captured features and reduce training time. Finally, to optimize the model’s performance, we use grid search to optimize the model’s parameters.
4 Materials and Methods
This section includes detailed explanations of the datasets used in this study as well as a variety of techniques, which are divided into two main categories: deep learning and data processing techniques.
4.1 Datasets
Six different datasets were used in this study: CVC-ClinicDB [24], CVC-ColonDB [25], ETIS-LaribPolypDB [26], Kvasir-SEG [27], and Kvasir-Sessile [27] with numbers of images of 612, 380, 196, 1000, and 196 respectively, as shown in Table 2 which shows each dataset with the corresponding number of images and masks, then the total number of them. We added 1500 normal images from HyperKvasir [28] to make the model able to differentiate between images that contain polyps and images without polyps, as shown in Table 3 which shows the number of images and masks of each dataset after adding normal images. Finally, to balance the dataset, active learning was used to annotate an additional 884 images from the unlabeled data found in the HyperKvasir dataset, which contains 99,417 unlabeled images. Images were added to the normal images to make 2384 total, and the dataset was named the Comprehensive Polyp Dataset (CPD) as shown in Table 4 which shows the number of images and masks after adding the labeled images to the normal images. Cost-sensitive uncertainty sampling was used as the labeling selection method. This method selects samples that are model uncertain and cheap to label.
4.2 Methods
This section is divided into two subsections: the first provides a summary of various deep learning models, and the second discusses various preprocessing techniques, including conditional random fields, locally shared features, data augmentation, and active learning.
4.2.1 Deep Learning Techniques
In this section, a brief introduction to the UNet, UNet++, ResUNet, and ResUNet++ models and the mechanisms behind those models, such as the attention mechanism, is provided.
4.2.2 UNet
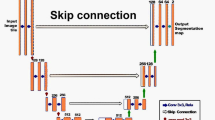
Olaf Ronneberger et al. developed UNet in 2015 at the University of Freiburg, Germany, for image segmentation in the medical field. Segmenting biomedical images is considered one of the most commonly used methods for any semantic segmentation task [29]. It is a fully convolutional neural network trained with fewer training samples. UNet is a U-shaped encoder–decoder network architecture consisting of four encoder and four decoder blocks linked by a bridge, as shown in Fig. 2 which shows the whole architecture of UNet. The encoder network has half the spatial dimensions and twice the number of filters for every encoder block. Likewise, the decoder network doubles the spatial dimensions while lowering the number of feature channels by half, then comes a convolution after non-linear activation function called ReLU, which helps the network learn different complex patterns in data, and its function can be denoted by:
where \(x=\) an input value
From the above function, the output of the activation function is the largest value between zero and the input data value. The output is a positive number when the value of the input data is greater than or equal to zero, while the output will be zero when the input data value is less than zero, so we can reformat Eq. 1 as shown below in Eq. 2:
where \(x=\) an input value
There is also a max-pooling function that is applied to the generated feature map to reduce its dimension and the amount of computation power carried over the network. The max-pooling output shape can be calculated as follows in Eq. 3:
where \(I_x\) is the input x, P is the pooling window, S is the stride, and the floor operation is applied on the numerator and dominator.
Architecture of UNet [30]
4.2.3 UNet++
The UNet++ is a redesign of the UNet in different aspects. The first aspect is the existence of convolutional layers on skip pathways, which improve gradient flow and reduce the semantic gap between encoder and decoder [31]. UNet remodelling helps in achieving high performance by applying deep supervision.
4.2.4 ResUNet
ResUNet is an abbreviation for Deep Residual UNet, which is based on encoder and decoder structures [32] and [33]. With fewer parameters and the help of a fully convolutional neural network, it can achieve high performance and good results in addition to the richness of skip connections that help transfer information between layers easily, which is a good application for polyp segmentation.
4.2.5 ResUNet++
The Deep Residual UNet architecture serves as the foundation for the ResUNet++ architecture. ResUNet++ is built with blocks such as attention [34], excitation and squeeze, atrous spatial pyramidal pooling [35, 36], and residual blocks. These blocks help in building a deeper neural network, enhance the cross-functionality between different channels, and minimize the computational cost. Since encoders and decoders have a problem with the complete sequence of information, the attention mechanism focuses on the most important attributes of the input sequence for each output. The attention mechanism can be generalized and calculated using Eq. 4:
Attention is the weighted sum of the values depending on the requested queries and the pair of keys, where q is the requested query for a set of two keys (K,V).
4.2.6 Data Processing Techniques
An overview of the data processing techniques used in this study and other studies. Techniques are classified into two techniques: post- and pre-processing techniques such as Conditional Random Fields and Locally Shared Features.
4.2.7 Conditional Random Fields
CRF, or Conditional Random Fields, is a post-processing tool that is frequently used to enhance the performance of algorithms, especially in image segmentation tasks [37]. A layer of CRF neural network is added in the form of a Recurrent Neural Network (RNN) in addition to UNET [38]. CRF helps solve the problem of mispredicted pixels and misclassified pixels. Debesh Jha et al. [18] conducted a comprehensive study to predict polyps by integrating CRF to enhance the model’s performance and be able to extract the most important features representing polyps, which in turn will enhance the overall results. CRF is represented by the Gibbs distribution as in the following Eq. 5:
where \(X_i\) is a random variable at i, \(E\left( X_i\right) \) is the energy function, and \(Z_j\) is the partition function.
4.2.8 Locally Shared Features
Yang et al. [39] introduced Locally Shared Features (LSF), which are better than the CRF. The technique is built on the basics of CRF. The aim of LSF is to improve the features of each image pixel by capturing the features of its neighbours. LSF is implemented in two steps: shifting and concatenating. In shifting, the feature map is shuffled in four directions (right, left, up, and down), while in concatenating, the original feature map is linked with the neighbouring feature map. LSF solved problems such as the computational problem of the CRF since it is time-consuming in addition to enhancing the model’s performance and enriching the image segmentation task through the complete Algorithm 1. LSF can be calculated using Eq. 6:
\(E\left( y\right) \) is the sum of applying LSF to pixel-wise classification, where \(x_i\) represents features and \(y_i\) is the pixel label.

4.2.9 Data Augmentation
Due to the shortage of available medical data [40], there are numerous techniques where the amount of data is artificially increased by generating a new data point from the residing data. These techniques are center cropping, random cropping, horizontal flip, vertical flip, random rotation [41], and scale augmentation. Data augmentation improves the model’s prediction accuracy [42] by increasing the generalization of the model, solving the problem of an unbalanced dataset and adding more data to the training set. Most of the recent studies used data augmentation to increase the polyp samples, either through preprocessing augmentation or test time augmentation, which is applied to the test dataset to boost the model’s performance and reduce overfitting problems [43, 44].
4.2.10 Active Learning
Active learning is a type of semi-supervised learning technique in which an algorithm can start questioning a user on the fly to give new labels for samples [45]. The goal of interactive learning is to achieve high accuracy with as few labeled samples as possible. Supervised segmentation algorithms, on the other hand, use previous knowledge from training samples as the ground truth. By incorporating deep convolutional neural networks in active learning and implementing a cost-effective algorithm to select samples that contributes to enhancing the segmentation task with fewer manual annotations, this method can produce a challenging classifier with optimized feature representation. M. Gorriz et al. [46] used the active learning approach for semantic segmentation of lesion areas in medical images. Since the existing deep learning models need a large number of labeled training samples, this could be a problem due to the limitations of medical data. Implementing such Cost-Effective Active Deep Learning (CEAL) reduced the time and cost of manual annotations [47,48,49].
5 Proposed Model for Segmenting Polyps
The proposed model aims to improve the accuracy of detecting adenomas by developing a medical system that can differentiate between healthy and unhealthy images. This is achieved by applying various image processing techniques to a large and diverse dataset of images. Additionally, the architecture of the existing deep learning model was modified, LSF was integrated, and new variations, such as UNet++, were implemented. Furthermore, grid search technique was employed to optimize all models. In this study, deep learning models like UNet, UNet++, ResUNet, and ResUNet++ were implemented. The proposed model consists of three main phases and two sub-phases: data fusion, including the active learning module as a sub-phase; preprocessing; training and testing (segmentation phase), including the hyperparameter tuning sub-phase, as shown in Figs. 3, 4, and 5.
5.1 Data Fusion Phase
The first phase of the proposed model is data fusion. As mentioned earlier, a full dataset was constructed and named the CPD, which contains a total of 9536 images and masks is the fusion of six datasets. To balance the dataset, CEAL is applied; thus, 884 extra images from the unlabeled data were added to build the normal images dataset, as shown in Fig. 3 where it depicts the data fusion and preprocessing phases.
5.2 Active Learning Module Sub-phase
The active learning module’s detailed flow, which is used in the data fusion phase, is shown in Fig. 5. The user module is a part of the active learning module, and it involves the interaction between the patient and the doctor during an endoscopic examination, which produces the images that are used to create the gastrointestinal dataset. UNet was used as a form of convolutional neural network to train and predict the data. The algorithm’s methodology depends on samples. Minority samples (most informative images) with low prediction confidence are one type of sample; these samples are the most uncertain and have the lowest prediction scores. We requested the assistance of two professional experts with extensive experience in the field. The first expert helped us label some ambiguous minority samples. To validate the newly labeled samples, we randomly select a subset and have them labeled by a second expert. Finally, the labeling results of the two experts were compared in order to demonstrate consistency and validate the labeled samples. Another form of sample is the majority of samples (most clearly classified images) that have high prediction scores. Because these samples’ predictions are so confident, the algorithm automatically adds labels without any burden on the human. Finally, minority and majority samples are added to the training set in an iterative manner.
5.3 Preprocessing Phase
In the preprocessing phase, all images were resized to 512 \(\times \) 512 after different iterations to determine the best size to keep image features, and then gaussian filters were applied to reduce Gaussian noise in the input images. Data augmentation functions were applied, like center and random cropping, rotating images, vertical and horizontal flip, and scale augmentation, to enhance the model’s performance and minimize the risk of overfitting, besides increasing the training sample. Finally, preprocessed and augmented images were added to the tensor flow dataset to be easily used as a high-performance input pipeline to the model.
5.4 Training and Testing Phase
To improve the performance of the four models (UNET, ResUNET, ResUNET++, and UNET++), we propose putting into practice a novel approach that makes use of training images and locally shared features. By utilizing both models’ complementing characteristics, this approach seeks to enhance segmentation abilities. In order to encode rich data from several perspectives, the integration procedure entails collecting intermediate characteristics from the encoder of each model. These locally shared features provide a more thorough interpretation of the input data by capturing both high-level contextual information and low-level details. Using locally shared features, we expect to significantly increase segmentation accuracy and generalization across a variety of datasets, furthering the state-of-the-art in medical image segmentation and related applications. Figure 4 illustrates the flow of the training and testing phases, which show the comparison between models after integrating LSF and the prediction results of the models after hyper-parameter tuning.
5.5 Hyper-Parameter Tuning Sub-phase
To be able to obtain the best performance and efficiency, we concentrate on optimizing several important parameters when hyperparameter tuning the four models (UNET, ResUNET, ResUNET++, and UNET++). Selecting suitable optimizers is the initial set of hyperparameters. To determine which one best fits each model’s design and convergence behavior, we investigate a variety of selections, including Adam, RMSprop, and SGD. The filter size is then addressed, with the goal of optimizing the convolutional layers’ receptive field to efficiently collect important image features. To balance feature representation and computational cost, we experiment with different filter dimensions. We also adjust the batch size while taking into account the trade-off between memory usage and training speed. While bigger batch sizes may speed up convergence but use more memory, smaller batch sizes may result in periodic updates but may be computationally consuming. Finally, for the purpose of maximizing spatial down-sampling while keeping important information, we investigate several pooling layer topologies, including max pooling and average pooling. During this hyperparameter tuning phase, we use methods such as grid search and compare performance across several metrics to find the ideal hyperparameter combinations that improve the segmentation accuracy and generalization abilities of each model.
6 Experiment Results
Many experiments were carried out to be able to identify the best parameters for the techniques implemented in the model. The study’s primary objective was to enhance the process of polyp segmentation to help the endoscopist identify the polyps more accurately, which can be reflected in reducing false-positive rates of polyp discovery by increasing precision and the dice coefficient. Two main experimental approaches are discussed in this section. In the first approach, deep learning models are used to apply the ablation study to the CPD dataset, while the second approach is to apply these models to each dataset (CVC-ClinicDB, CVC-ColonDB, ETIS LaribPolyp, KVASIR-SEG, Kvasir-Sessile, and CPD). The results of the ablation study and those obtained using UNet, UNet++, ResUNet, and ResUNet++ with LSF are then presented.
6.1 Experimental Settings
The LSF technique was used before running the model, and the data was split into 80% training and 20% testing. The predicted images were investigated with the masks to assess the performance of the proposed techniques by contrasting their DSC, recall, and precision. The proposed PolySeg Plus model was trained on a system equipped with an 11th Gen Intel (R) Core (TM) i7-11800H @ 2.30 GHz processor, 16 GB of RAM, a NVIDIA GeForce RTX 3060 GPU, and 1 TB of SSD storage, in addition to some experiments conducted on Google Colab Pro. All the experiments were performed using Anaconda version 2.0 and Python 3.7. Based on the following ablation study, the hyperparameters used were batch size of 64, a filter size of 5 \(\times \) 5, max pooling layer. The optimizers used were Adam, SGD, and Nadam.
6.2 Evaluation Metrics
The performances of different model variations are measured by three commonly used metrics: Dice Similarity Coefficient (DSC), Recall and Precision. DSC is used to measure the similarity between two sets of data, and it is widely used to evaluate the output of the image segmentation operation. DSC is defined by the following formula, as shown in Eq. 7:
where M is the predicted set of image pixels and N is the image ground truth, and DSC is twice the overlapped area between M and N divided by the number of pixels in each image. Recall measures the completeness of the model in detecting the number of captured positive samples, whereas precision measures how many of these positive samples match the image ground truth. Recall and precision are defined by Eqs. 8 and 9, respectively.
In this study, TP means true positive (presence of polyp in image), FP means false positive (predicted polyp in the image, but the image does not contain polyp), and FN means false negative (the image contains polyp but is predicted to be without polyp).
6.3 Ablation Study
An ablation study in deep learning is a method used to understand the impact of individual components or elements of a neural network on its overall performance. This is typically done by removing or “ablating” one or more parts of the network and evaluating the effect on the model’s accuracy or other performance metrics. This study can help identify the most important features or components of the network, as well as potential areas for improvement. It can also be used to compare different architectures or configurations of a network. This study is applied in three parts: active learning, preprocessing, and hyperparameter tuning, All three parts are applied to four different deep learning models: UNet, UNet++, ResUNet, and ResUNet++ on the CPD dataset. For evaluation, image DSC is used to evaluate the performance of the tested models.
6.3.1 Active Learning Method
In the CEAL, various factors can impact the accuracy of the model’s prediction, such as the number of iterations carried out in the active learning process, the quantity of predictions made in each iteration, the number of samples with the greatest uncertainty selected to be included in the training set per iteration, and the size of each batch while training.
6.3.2 Case Study 1: Altering the Number of Active Learning Iterations and the Number of Predictions for Each Iteration
Table 5 displays the outcomes of four attempts where a UNet model was trained on a Hyper Kvsair dataset. Each experiment used a variable number of iterations and different predictions for each iteration. The number of epochs multiplied by the length of time needed to complete one training iteration is shown in the “Epoch Training Time” column. The “DSC” column displays the Dice Similarity Coefficient, a metric used to assess the model’s performance, with higher values indicating better performance. The model’s performance (DSC) greatly increased with both the number of iterations and the number of predictions per iteration. The highest accuracy of 0.8887 was achieved by trial 4, which took 40 s to complete 60 epochs. Our findings indicate that having twice as many predictions as iterations leads to the best results. Additionally, starting the training with a higher number of uncertainty samples can impact the accuracy of the model, and the optimal number was found to be five samples labeled by a human.
6.3.3 Case Study 2: Altering Batch Size
Results from various trials in the experiment on active learning are shown in Table 6. It demonstrates the effects of various batch sizes and training durations on the Dice Similarity Coefficient (DSC) performance parameter. A batch size of 32 and a training time of 20 epochs, lasting 26 s each, resulted in the highest DSC of 0.8524. However, the batch size of 128 did not perform as well, with higher epoch numbers and longer training times compared to the batch size of 32. Therefore, we chose a batch size of 32 for further analysis.
6.3.4 Preprocessing
Image processing before training is crucial, as it pre-processes and enhances the quality of the images used for training, resulting in improved accuracy and performance of the model. It includes techniques such as resizing, normalization, and noise reduction, which help reduce variability and make the images more consistent, leading to a more robust model. Image processing also helps in removing any irrelevant information and increasing the signal-to-noise ratio, making it easier for the model to learn and recognize patterns in the data. Therefore, image processing is a crucial step in ensuring the success of the machine learning model and its ability to accurately perform its intended task.
6.3.5 Case Study 1: Applying Gaussian Filters
Gaussian filters are commonly used in image processing to remove Gaussian noise and smooth out images. The importance of Gaussian filters lies in their ability to enhance the quality of images, improve the accuracy of computer vision algorithms, and facilitate the process of image analysis. For several image segmentation models, performance measures in terms of dice are shown in Table 7. With and without Gaussian filters are used to compare the models in these two conditions. The results show that the models with Gaussian filters perform better than those without them in general, with ResUNet++ getting the best score of 0.8812 in the condition when Gaussian filters were used. Algorithm 2 demonstrates the process of applying a Gaussian filter to the input images.

6.3.6 Case Study 2: Applying LSF
After applying the Gaussian filters to the training images, LSF is applied directly before training. Locally shared features in deep learning refer to the similarities between the features learned in different layers of a deep neural network. These locally shared features are crucial because they allow the network to learn common representations from the input data, which in turn enhances the overall accuracy of the model. They also help reduce the number of parameters and computational complexity in the network, making it more efficient and easier to train. Additionally, the locally shared features promote generalization, as the network can recognize similar patterns in the data even when it is presented in different forms. Overall, locally shared features play a vital role in the success of deep learning models. Table 8 contrasts the effectiveness of several image segmentation models using the Locally Shared Features (LSF) and without the Locally Shared Features (LSF) approache. UNet, UNet++, ResUNet, and ResUNet++ are among the models. Values for each model and method’s relevant evaluation measure, such as DSC, show that Locally Shared Features often enhance model performance. LSF did not enhance much in UNet; however, LSF achieved a great improvement with ResUNet++, with an accuracy of 0.9312.
6.3.7 Hyperparameter Tuning
To find the best structure and settings for a CNN model, it is necessary to take into account the type of task and any possible difficulties related to it. The goal of an ablation study is to gain a clear understanding of how the model performs by examining the effects of changing certain parts. By making changes to various components or parameters of the model, variations in performance can be observed. This approach allows for the identification of any potential declines in performance, which can then be corrected by adjusting and fine-tuning the network. As a result, we have experimented with our base CNN model multiple times by changing the number of layers, filter sizes, filter numbers, parameters, and other variables, to attain optimal performance with minimal computational resources. Hyperparameter optimization was done using the GridSearchCV library in scikit-learn to be able to choose the best parameters.
6.3.8 Case Study 1: Altering Optimizer
Optimizers are crucial components in Convolutional Neural Networks (CNNs) as they control the model’s learning process. They determine how the model updates its parameters based on the loss function and training data. The optimizer determines the speed and direction of learning and helps the model reach its optimal accuracy. Without the optimizer, the model’s learning process would be slow, unstable, and possibly converge to a suboptimal solution. Hence, the choice of optimizer is important as it can greatly affect the overall performance of the CNN. Different optimization algorithms, including Adam, Nadam, and Stochastic Gradient Descent SGD were tested to determine the best optimizer. Table 9 shows the performance of various models (UNet, UNet++, ResUNet, and ResUNet++) using three distinct optimizers (Adam, Nadam, and SGD). The values in the table show the related evaluation metrics (DSC) that each model and each optimizer were able to accomplish. With the highest scores from the Nadam and SGD optimizers, ResUNet++ seems to be the model that performs the best overall. The optimal parameters for UNet, UNet++, ResUNet, and ResUNet++ were SGD, Adam, SGD, and Nadam. We choose the previous optimizers for each model for further ablation studies.
6.3.9 Case Study 2: Altering Filter Size
The size of filters in a Convolutional Neural Network (CNN) plays a crucial role in determining the network’s ability to learn useful features from the input data. Large filters can capture global patterns in the input, while small filters can capture local patterns. By varying the size of filters, a CNN can learn a hierarchy of features, from simple edge detection to more complex shapes and objects. This allows the network to make more informed decisions when classifying the input data. Additionally, using different sized filters can also reduce overfitting and improve the overall accuracy of the model. It is important to experiment with different filter sizes to find the optimal configuration for a specific problem and dataset. With different filter sizes (2 \(\times \) 2, 3 \(\times \) 3, 5 \(\times \) 5, and 7 \(\times \) 7), Table 10 compares the performance of various semantic segmentation models (UNet, UNet++, ResUNet, and ResUNet++). The DSC metric is used to assess the performance of the models, and the values represent the equivalent scores attained by each model using various filter sizes. With a maximum score of 0.9242, ResUNet++ performs most effectively overall among the models and filter sizes. For 2 \(\times \) 2 filter size, the highest accuracy is achieved by ResUNet++, with an accuracy of 0.8843; except for the ResUNet, the accuracy was lowered by 3.23%. However, the filter size 3 \(\times \) 3 did slightly enhance the overall performance for all models. The 5 \(\times \) 5 filter improved the results of the models using the previous filter 3 \(\times \) 3 by an average of 3.16%. As shown in Table 10, filter size 7 \(\times \) 7 achieved the highest accuracy for all models: UNet, UNet++, ResUNet, and ResUNet++, with accuracies of 0.8826, 0.9137, 0.9000, and 0.9387. As a result, the 7 \(\times \) 7 filter size is selected for each model for further ablation research.
6.3.10 Case Study 3: Altering Batch Size
The batch size refers to the number of images utilized in each iteration during the training process of the model. Having a larger batch size can result in longer convergence times, but a smaller batch size may affect the model’s performance negatively. The complexity of medical images can also impact the model’s performance when using different batch sizes, leading to varying results. At various batch sizes (16, 32, 64, and 128), Table 11 compares the performance of various models (UNet, UNet++, ResUNet, and ResUNet++). According to the DSC metric, the models are assessed. ResUNet++ outperformed other models in the comparison, achieving the greatest results overall across a range of batch sizes. We discovered that batch sizes of 32 and 64 achieved the highest average accuracy of 0.8836 and 0.8886, respectively, as shown in Table 11. The highest DSC accuracy for 32 and 64 batch sizes was 0.9281 and 0.9321 for ResUNet++, respectively. As a result, 64 batch sizes were used for further ablation research. According to literature studies, larger batch sizes can result in favorable outcomes and improved generalization when selecting network optimizers [50] and figuring out the model learning rate [51]. The best way to choose the best parameters is to experiment with different batch sizes while controlling other factors, then compare the results.
6.3.11 Case Study 4: Altering the Pooling Layer’s Configuration
Pooling layers is an important component of convolutional neural networks (CNNs) used in deep learning for image processing tasks. The primary purpose of pooling layers is to down-sample the spatial dimensions of the feature maps generated by convolutional layers. This downsampling helps reduce the number of parameters in the model, which in turn can help prevent overfitting, reduce computational complexity, and improve model efficiency. Pooling layers can also help the model be invariant to small translations, rotations, and distortions in the input image, thereby increasing its ability to generalize to new, unseen data. The performance of multiple models using different pooling layers (Max, Average, and Global) is compared in Table 12 using DSC evaluation measures. The models are divided into ResUNet, ResUNet++, UNet, and UNet++. Over all pooling layers, ResUNet++ outperformed the other models and achieved the highest accuracy scores. The highest accuracy was recorded for the max pooling layer for all models, while global pooling did not enhance the model’s performance and the lowest accuracy was 0.8322 for the UNet model. The max pooling layer is therefore chosen for further ablation studies. We also observed that the performance of the model degraded upon increasing the number of epochs above 60, so the optimal number of epochs was between 50 and 60 epochs.
Based on the previous ablation study, the entire structure of the CNN model is modified, and the outcomes are documented. This process is carried out for every example under consideration. In the active learning part, two case studies were carried out. In the first case study, we found that the optimal number of iterations is 30, and the number of predictions per iteration is 60, while in the second case, using a batch size of 32 achieved the highest performance. In the preprocessing, we experimented with applying gaussian filters and LSF. Gaussian filters enhanced the average results by 5.68%; also, using LSF techniques enriched the performance of the model, and the results were compared before and after using the LSF. The last part is the hyperparameter tuning part, and based on different experiments, the following parameters were chosen: The best optimizers, SGD, Adam, SGD, and Nadam, were chosen for the following models: UNet, UNet++, ResUNet, and ResUNet++, respectively; a filter size of 5 \(\times \) 5, batch size of 64; and max pooling layer. The results of the entire ablation study are presented in Tables 5 and 6 for active learning and Tables 7 and 8 for preprocessing, while Tables 9, 10, 11, and 12 contain all the results related to the model’s hyperparameter tuning.
6.3.12 Hyperparamter Tuning Using Grid Search
In this experiment, we carried out a thorough hyperparameter tuning analysis for four deep learning models, UNet, UNet++, ResUNet, and ResUNet++, which are frequently employed in medical image segmentation tasks. A key step in machine learning is hyperparameter tuning, which improves the generalization and performance of the models. The model’s capacity to precisely identify regions of interest in medical images is substantially impacted by the hyperparameters that were chosen. With regard to the Dice Similarity Coefficient (DSC) measure, we specifically investigated the effects of several optimizers (Adam, Nadam, and SGD), filter sizes (2 \(\times \) 2, 3 \(\times \) 3, 5 \(\times \) 5, and 7 \(\times \) 7), batch sizes (16, 32, 64, and 128), and pooling layer settings (Max, Average, and Global). Table 13 displays the outcomes of the hyperparameter tuning experiment for the four models UNet, UNet++, ResUNet, and ResUNet++. Each row represents a particular model configuration, which includes the optimizer selected, the filter size, batch size, pooling layer, and the DSC as an evaluation metric. It is clear from the findings that hyperparameter tuning has had a considerable impact on the models’ performance. Comparing the original UNet and ResUNet architectures to the UNet++ and ResUNet++ models, significantly higher DSC ratings were obtained. Additionally, both the ResUNet and ResUNet++ models performed better when they were tuned using the SGD optimizer with a higher filter size of 7 \(\times \) 7 and a batch size of 64. The Adam optimizer, a filter size of 3 \(\times \) 3, and a reduced batch size of 32, on the other hand, improved the performance of the UNet and UNet++ models. Additionally, for all models, the Max pooling layer consistently produced positive outcomes. In general, it has been found that improving the segmentation accuracy of the models through hyperparameter tuning is successful. With the SGD optimizer, a 7 \(\times \) 7 filter size, a batch size of 64, and an average pooling layer, the ResUNet++ model stands out as the best-performing architecture. It achieved a DSC score of 0.9611, which is outstanding. The full details of Grid Search are displayed in Algorithm 3.

6.4 Experimental Results Analogy of Baseline Models and PolySeg Plus on CVC-ClinicDB Dataset
The CVC-ClinicDB dataset contains 612 images. After applying LSF to ResUNet, ResUNet++, UNet, and UNet++, there was a great improvement in the performance of all algorithms. In terms of DSC and precision, ResUNet++ + LSF improved by 3.55% and 7.65%, respectively, compared to ResUNet++ + CRF. UNet++ + LSF achieved a DSC of 0.7511, as shown in Table 14 where the results of our model are compared to the studies made by Ronneberger et al. [30], Zhang et al. [52], and Debesh Jha et al. [18].
6.5 Experimental Results Analogy of Baseline Models and PolySeg Plus on CVC-ColonDB Dataset
In CVC-ColonDB, the number of images is 380, which is nearly half the number of images in CVC-ClinicDB. After applying the ResUNet++ + LSF (PolySeg Plus model variation), there was a slight improvement in DSC and recall compared to the baseline model (ResUNet++ + TTA), as shown in Table 15 where the results of our model are compared to the study made by Debesh Jha et al. [18], also; hence, we do not have to apply test time augmentation as applied in the baseline model.
6.6 Experimental Results Analogy of Baseline Models and PolySeg Plus on ETIS Larib Polyp DB Dataset
Comparing the results of the models on the other datasets, the results were not significant since the number of images in the datasets is 196, which is relatively small. However, we applied different variations of PolySeg Plus to test the ability of models on a small dataset. As shown in Table 16 where the results of our model are compared to the study made by Debesh Jha et al. [53], applying LSF to ResUNet++ improved DSC by about 11.83% while improving precision and recall by 19.2% and 18.45%, respectively.
6.7 Experimental Results Analogy of Baseline Models and PolySeg Plus on KVASIR-SEG Dataset
Kvasir-seg has the largest number of images and masks (totaling 1000 images) compared to other datasets. UNet++ and LSF performed the best compared to UNet+ LSF and the baseline model UNet; the average DSC and precision of the two PolySeg Plus variation models were 0.8207 and 0.8250, respectively, as shown in Table 17 where the results of our model are compared by the studies made by Ronneberger et al. [30] and Debesh Jha et al. [18].
6.8 Experimental Results Analogy of Baseline Models and PolySeg Plus on Kvasir-Sessile Dataset
The baseline model was implemented using ResUNet++ + TTA, while in PolySeg Plus using the LSF technique, results were enhanced in terms of DSC, and the gap between the recall and precision was reduced by 7.83%, as shown in Table 18 where the results of our model are compared by the studies made by Debesh Jha et al. [18]. The results were not significant since the Kvasir-Sessile dataset has a small number of images.
6.9 Experimental Results of Poly-Seg Plus on Comprehinsive Polyp Dataset
For generalization and enhancing model performance on different image variations, PolySeg Plus was implemented, a comprehensive polyp dataset that fuses CVC-ClinicDB, CVC-ColonDB, ETIS Larib Polyp, Kvasir-Seg, Kvasir-Sessile, and normal images. ResUNet++ + LSF achieved the highest performance compared to ResUNet, UNet, and UNet++. ResUNet++ + LSF achieved a DSC of 0.9547, as shown in Table 19 and Fig. 7, which is higher than the average DSC of the other individual datasets, which is 0.8310, as shown in Fig. 6 where the results of the polySeg Plus are compared to the study by Debesh Jha et al. [18].
7 Results Discussion
The PolySeg Plus has introduced a new high-performance model variation that achieved higher results than the existing baseline models, which was attributed to ResUNet++ + LSF. Three major factors contributed to the significant results. To begin, good data preprocessing entails resizing the image size to a fixed size and applying Gaussian filters to eliminate Gaussian noise and improve the quality of the images to retain the majority of their features. Second, applying the LSF technique, which is an enhancement of the CRF, to extract the most important features. Finally, the hyperparameter tuning can make significant improvements in the performance of the model by altering different parameters. For both the PolySeg Plus and baseline models, the average results of ResUNet++ and UNet were compared. There is an enhancement in DSC and recall by 8.29% and 8.06% using ResUNet++ on the datasets CVC-ClinicDB, CVC-ColonDB, ETIS Larib Polyp, KVASIR-SEG, and Kvasir-Sessile, respectively. On the other hand, UNet had an enhancement in DSC and recall of 7.35% and 7.19% on the datasets CVC-ClinicDB and KVASIR-SEG as shown in Fig. 6, and the results are verified qualitatively, as shown in Fig. 8 where the output segmented image of the PolySeg Plus is compared to the mask on the CPD dataset, while Fig. 9 shows the visualization of segmented images and the corresponding mask results of our method compared to baseline models. Finally, Figs. 10 and 11 present the train and validation progress of PolySeg Plus models on CPD through epochs in terms of DSC respectively. As we can see, the proposed model shows a better segmentation result on different sizes of polyps, particularly small and flat polyps, which the other baseline models, UNet and ResUNet, failed to detect. Furthermore, it has been observed that the baseline models in some images are inaccurate by segmenting polyps that do not exist, which increases the number of false positive samples and reduces the accuracy and effectiveness of the model, potentially resulting in harmful and costly medical actions or interventions for the patient.
Comparing the results of PolySeg Plus with the baseline paper [18] on the datasets CVC-ClinicDB and KVASIR-SEG
7.1 Previous Work Results Discussion
A summarised Table 20 compares previous works on each dataset to Poly-Seg Plus in terms of dice similarity coefficient (DSC), mean DSC, and accuracy. the highest DSC of 0.9558, achieved by PolySeg Plus on CVC-CLINICDB, when compared to other studies on the same dataset, which show 3.55%, 11.68%, 3.98%, and 0.99% improvement on SOTA [18,19,20, 23], respectively. Regarding CVC-COLONDB, PolySeg Plus achieved a DSC of 0.8947 and an improvement on SOTA [18, 20] of 4.73% and 9.07%, respectively. Comparing results on ETIS LARIB POLYP DB, our proposed model improved the result by 11.83% compared to SOTA [18]. Investigating results on the KVASIR-SEG dataset, PolySeg Plus achieved a DSC of 0.9476 and higher results than other SOTA [18, 20, 22] by 9.68%, 3.26% and 9.61%, respectively. Finally, there was a great enhancement of 9.81% on KVASIR-SESSILE compared to SOTA [18]. In our opinion, this improvement has helped to overcome the shortcomings in earlier studies, such as the small number of images and the deteriorated image quality caused by image resizing, which contributes to the loss of significant image features.
8 Hypothesis and Limitations
This section is a crucial part of the study since it explores the essential presumptions and expectations that support the use of deep learning models with locally shared information for polyp segmentation. This section outlines our method to achieving more precise and reliable polyp segmentation as well as the basic idea that underlies our research. Furthermore, we explore potential problems that can impair the generalizability, dependability, and applicability of our suggested solution while being open about the methodology’s inherent limits. This section contributes to a thorough and detailed understanding of the implications and future prospects for polyp segmentation using deep learning algorithms by examining both the potential and the limits of our study.
8.1 Hypothesis
The main objective of this research is to determine whether using deep learning models with locally shared features, such as UNet, UNet++, ResUNet, and ResUNet++, can effectively segment polyps in medical imaging data. In comparison to conventional techniques, we believe that the inclusion of locally shared features inside these state-of-the-art structures will improve segmentation performance.
Early detection and appropriate treatment of gastrointestinal problems depend heavily on the detection and segmentation of polyps in medical imaging, especially in endoscopy and colonoscopy. Convolutional neural networks (CNNs), in particular, have shown incredible performance in a variety of image segmentation tasks. However, successful segmentation is significantly hampered by the intricate and erratic forms of polyps.
In this study, we start by implementing the standard UNet architecture, which has been successful in medical image segmentation tasks. To develop the UNet++ model, we then extend the UNet to include locally shared features. The ResUNet++ model is the outcome of our enhancements to the ResUNet architecture, which is well-known for its skip connections. We carried out experiments on a large and varied collection of polyp images acquired from several health care providers to verify our hypothesis. The dataset has undergone preprocessing to standardize image resolution and remove any noise or artifacts. On this dataset, we use optimal hyperparameters to refine the deep learning models through a rigorous training process. Figure 12 shows the complete Flowchart of PolySeg Plus model.
The following outcomes are what we anticipate will happen as a result of the inclusion of locally shared features in the UNet, UNet++, ResUNet, and ResUNet++ architectures:
-
1.
Improved segmentation accuracy: It is believed that the locally shared features would provide better contextual information, allowing the models to more accurately determine polyp boundaries, particularly in areas with complex structures and low contrast.
-
2.
Reduced Overfitting: The models may generalize to unseen polyp images more effectively as a result of the added contextual information from locally shared features, which lowers the potential risk of overfitting.
-
3.
Possibility of Real-Time Application: These architectures’ efficiency and processing advantages may make it possible to segment polyps in real-time during endoscopic surgeries.
-
4.
Robustness to Different Polyp Shapes and Sizes: The locally shared features are anticipated to enhance the models’ capacity to manage polyps of various shapes, sizes, and orientations by capturing fine-grained spatial details.
8.2 Limitations
Deep learning models sometimes require significant computer resources and training time, particularly those with locally shared features. It may be computationally demanding to train these models on large datasets of high-resolution medical pictures, and access to robust hardware, such as top-tier GPUs or TPUs, may be necessary. This drawback might prevent our suggested solution from being widely used in environments with limited resources. In addition, Small-sized polyp detection and precise segmentation continue to be challenging, especially when the polyps are covered up by background noise or other anatomical structures. Our models may still have some trouble accurately identifying and distinguishing tiny polyps despite the inclusion of locally shared features.
While our proposed approach shows promising results in the experimental setting, its clinical applicability and impact need to be validated through rigorous clinical trials and evaluations. Real-world medical environments may present additional complexities and variations that could affect the performance of the models. Although the experimental results from our suggested approach are encouraging, extensive clinical studies and evaluations are still required to confirm its clinical applicability and impact. Additional complications and variables that may be present in real-world medical settings could impair the performance of the models.
9 Conclusion
In this paper, different models of neural networks and deep learning models for semantic polyp segmentation were applied. We carried out the study on six datasets, where five datasets contained unhealthy images (abnormal dataset) that had polyps and the other dataset contained healthy images (normal dataset) that were free of polyps. Since the number of unhealthy images was greater than the number of labeled healthy images, the dataset was considered imbalanced. A cost effective active deep learning algorithm was applied to help label the healthy images with less cost to balance both classes: the healthy image class and the unhealthy image class. A full polyp dataset of different clinical images was established to increase the training data for better model performance, followed by reading both images and masks, applying a Gaussian filter to reduce Gaussian noise or blurriness in the input images, and finally applying data augmentation techniques to increase the training set. Several experiments were conducted, and the results have shown that applying ResUNet++ + LSF to each dataset and the new dataset that contains more training samples helps a lot, along with data augmentation. Also, UNet++ was introduced, which showed better performance than UNet. To enhance the model’s results, hyperparameter tuning was applied using Grid Search to find the best possible parameter combinations for each model. The primary goal of this study was to develop a robust semantic segmentation model that has significant generalization ability and can be implemented in the medical field. We believe that enhancing the recall in such a model will help endoscopists be able to recognize polyps easily if they are not clear during examination and will add value to the domain of colonoscopy.
Data Availability
All datasets are accessible via the internet.
References
Xi, Y., Xu, P.: Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 14, 101174 (2021). https://doi.org/10.1016/j.tranon.2021.101174
Saad, A.I., Omar, Y.M., Maghraby, F.A.: Predicting drug interaction with adenosine receptors using machine learning and smote techniques. IEEE Access 7, 146953–146963 (2019). https://doi.org/10.1109/ACCESS.2019.2946314
Miller, K.D., et al.: Cancer treatment and survivorship statistics, 2022. CA Cancer J. Clin. 72, 409–436 (2022). https://doi.org/10.3322/caac.21731
Soleimaninejad, M., Sharifian, M., et al.: Evaluation of colonoscopy data for colorectal polyps and associated histopathological findings. Ann. Med. Surg. 57, 7–10 (2020). https://doi.org/10.1016/j.amsu.2020.07.010
Ray-Offor, E., Jebbin, N.: Risk factors for inadequate bowel preparation during colonoscopy in Nigerian patients. Cureus (2021). https://doi.org/10.7759/cureus.17145
Mori, Y., et al.: Real-time use of artificial intelligence in identification of diminutive polyps during colonoscopy: a prospective study. Ann. Intern. Med. 169, 357–366 (2018). https://doi.org/10.7326/M18-0249
Barua, I., et al.: Real-time artificial intelligence-based optical diagnosis of neoplastic polyps during colonoscopy. NEJM Evidence 1, EVIDoa2200003 (2022). https://doi.org/10.1056/EVIDoa2200003
Reverberi, C., et al.: Experimental evidence of effective human-ai collaboration in medical decision-making. Sci. Rep. 12, 14952 (2022). https://doi.org/10.1038/s41598-022-18751-2
Choe, J., et al.: Content-based image retrieval by using deep learning for interstitial lung disease diagnosis with chest ct. Radiology 302, 187–197 (2022). https://doi.org/10.1148/radiol.2021204164
Naveen Kumar, G., Reddy, V.: In: High performance algorithm for content-based video retrieval using multiple features. https://doi.org/10.1007/978-981-19-0011-2_57
Tuladhar, S., Alsadoon, A., Prasad, P., Ali, A.E., Alrubaie, A.: A novel solution of deep learning for endoscopic ultrasound image segmentation: enhanced computer aided diagnosis of gastrointestinal stromal tumor. Multimed. Tools Appl. 81, 23845–23865 (2022). https://doi.org/10.1007/s11042-022-11936-x
Ouyang, C., et al.: Self-supervised learning for few-shot medical image segmentation. IEEE Trans. Med. Imaging 41, 1837–1848 (2022). https://doi.org/10.1109/TMI.2022.3150682
Guo, Q., Fang, X., Wang, L., Zhang, E.: Polyp segmentation of colonoscopy images by exploring the uncertain areas. IEEE Access 10, 52971–52981 (2022). https://doi.org/10.1109/ACCESS.2022.3175858
Yao, L., et al.: Scheme and dataset for evaluating computer-aided polyp detection system in colonoscopy, pp. 1–5. IEEE (2022). https://doi.org/10.1109/ISBI52829.2022.9761699
Suganyadevi, S., Seethalakshmi, V., Balasamy, K.: A review on deep learning in medical image analysis. Int. J. Multimed. Inf. Retriev. 11, 19–38 (2022). https://doi.org/10.1007/s13735-021-00218-1
Koo, E., Kim, G.: A hybrid prediction model integrating garch models with a distribution manipulation strategy based on lstm networks for stock market volatility. IEEE Access 10, 34743–34754 (2022). https://doi.org/10.1109/ACCESS.2022.3163723
Mandal, S., Chaudhuri, S.S.: Polyps segmentation using fuzzy thresholding in hsv color space, pp. 1–5. IEEE (2020). https://doi.org/10.1109/HYDCON48903.2020.9242852
Jha, D., et al.: A comprehensive study on colorectal polyp segmentation with resunet++, conditional random field and test-time augmentation. IEEE J. Biomed. Health Inform. 25, 2029–2040 (2021). https://doi.org/10.1109/JBHI.2021.3049304
Banik, D., Roy, K., Bhattacharjee, D., Nasipuri, M., Krejcar, O.: Polyp-net: a multimodel fusion network for polyp segmentation. IEEE Trans. Instrum. Meas. 70, 1–12 (2020). https://doi.org/10.1109/TIM.2020.3015607
Qiu, Z., et al.: Bdg-Net: Boundary Distribution Guided Network for Accurate Polyp Segmentation, vol. 12032, pp. 792–799. SPIE (2022). https://doi.org/10.1117/12.2606785
Mohapatra, S., Pati, G.K., Mishra, M., Swarnkar, T.: Upolyseg: a u-net-based polyp segmentation network using colonoscopy images. Gastroenterol. Insights 13, 264–274 (2022). https://doi.org/10.3390/gastroent13030027
Gautam, A., Das, S., Sharma, P., Maji, P., Balabantaray, B.K.: Sau-net: Scale Aware Polyp Segmentation Using Encoder–Decoder Network, pp. 1–5. IEEE (2022). https://doi.org/10.1109/TENSYMP54529.2022.9864338
Tran, S.-T., Nguyen, M.-H., Dang, H.-P., Nguyen, T.-T.: Automatic polyp segmentation using modified recurrent residual unet network. IEEE Access 10, 65951–65961 (2022). https://doi.org/10.1109/ACCESS.2022.3184773
Bernal, J., et al.: Wm-dova maps for accurate polyp highlighting in colonoscopy: validation vs. saliency maps from physicians. Comput. Med. Imaging Graph. 43, 99–111 (2015). https://doi.org/10.1016/j.compmedimag.2015.02.007
Bernal, J., Sánchez, J., Vilarino, F.: Towards automatic polyp detection with a polyp appearance model. Pattern Recogn. 45, 3166–3182 (2012). https://doi.org/10.1016/j.patcog.2012.03.002
Silva, J., Histace, A., Romain, O., Dray, X., Granado, B.: Toward embedded detection of polyps in wce images for early diagnosis of colorectal cancer. Int. J. Comput. Assist. Radiol. Surg. 9, 283–293 (2014). https://doi.org/10.1007/s11548-013-0926-3
Jha, D., et al.: Kvasir-seg: A Segmented Polyp Dataset. Springer, pp. 451–462 (2020). https://doi.org/10.1007/978-3-030-37734-2_37
Borgli, H., et al.: Hyperkvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci. Data 7, 283 (2020). https://doi.org/10.1038/s41597-020-00622-y
Liu, F., Wang, L.: Unet-based model for crack detection integrating visual explanations. Constr. Build. Mater. 322, 126265 (2022). https://doi.org/10.1016/j.conbuildmat.2021.126265
Ronneberger, O., Fischer, P., Brox, T.: U-net: Convolutional Networks for Biomedical Image Segmentation. Springer, pp. 234–241 (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Li, Z., Zhang, H., Li, Z., Ren, Z.: Residual-attention unet++: a nested residual-attention u-net for medical image segmentation. Appl. Sci. 12, 7149 (2022). https://doi.org/10.3390/app12147149
Weng, W., Zhu, X.: Inet: convolutional networks for biomedical image segmentation. IEEE Access 9, 16591–16603 (2021). https://doi.org/10.1109/ACCESS.2021.3053408
Sabir, M.W., et al.: Segmentation of liver tumor in ct scan using resu-net. Appl. Sci. 12, 8650 (2022). https://doi.org/10.3390/app12178650
Maji, D., Sigedar, P., Singh, M.: Attention res-unet with guided decoder for semantic segmentation of brain tumors. Biomed. Signal Process. Control 71, 103077 (2022). https://doi.org/10.1016/j.bspc.2021.103077
Xu, W., Liu, H., Wang, X., Qian, Y.: Liver Segmentation in ct Based on Resunet with 3d Probabilistic and Geometric Post Process, pp. 685–689. IEEE (2019). https://doi.org/10.1109/SIPROCESS.2019.8868690
Ibrahim, S., et al.: Lung Segmentation Using Resunet++ Powered by Variational Ato Encoder-Based Enhancement in Chest X-ray Images. Springer, pp. 339–356 (2022). https://doi.org/10.1007/978-3-031-12053-4_26
Chen, S., Gamechi, Z.S., Dubost, F., van Tulder, G., de Bruijne, M.: An end-to-end approach to segmentation in medical images with cnn and posterior-crf. Med. Image Anal. 76, 102311 (2022). https://doi.org/10.1016/j.media.2021.102311
Thanh, N.C., Long, T.Q., et al.: Crf-efficientunet: an improved unet framework for polyp segmentation in colonoscopy images with combined asymmetric loss function and crf-rnn layer. IEEE Access 9, 156987–157001 (2021). https://doi.org/10.1109/ACCESS.2021.3129480
Yang, Z., Yu, H., Sun, W., Mao, Z., Sun, M.: Locally shared features: an efficient alternative to conditional random field for semantic segmentation. IEEE Access 7, 2263–2272 (2018). https://doi.org/10.1109/ACCESS.2018.2886524
Lo, J., Cardinell, J., Costanzo, A., Sussman, D.: Medical augmentation (med-aug) for optimal data augmentation in medical deep learning networks. Sensors 21, 7018 (2021). https://doi.org/10.3390/s21217018
Ma, Y., Chen, X., Sun, B.: Polyp Detection in Colonoscopy Videos by Bootstrapping via Temporal Consistency, pp. 1360–1363. IEEE (2020). https://doi.org/10.1109/ISBI45749.2020.9098663
Jheng, Y.-C., et al.: A novel machine learning-based algorithm to identify and classify lesions and anatomical landmarks in colonoscopy images. Surg. Endosc. 36, 640–650 (2022). https://doi.org/10.1007/s00464-021-08331-2
Chen, H., Cao, P.: Deep Learning Based Data Augmentation and Classification for Limited Medical Data Learning, pp. 300–303. IEEE (2019). https://doi.org/10.1109/ICPICS47731.2019.8942411
Kebaili, A., Lapuyade-Lahorgue, J., Ruan, S.: Deep learning approaches for data augmentation in medical imaging: a review. J. Imaging 9, 81 (2023). https://doi.org/10.3390/app12178650
Zheng, Y., Gao, Y., Lu, S., Mosalam, K.M.: Multistage semisupervised active learning framework for crack identification, segmentation, and measurement of bridges. Comput.-Aid. Civ. Infrastruct. Eng. 37, 1089–1108 (2022). https://doi.org/10.1111/mice.12851
Gorriz, M., Carlier, A., Faure, E., Giro-i Nieto, X.: Cost-effective active learning for melanoma segmentation (2017). arXiv preprint arXiv:1711.09168. https://doi.org/10.48550/arXiv.1711.09168
Rawat, S., et al.: How useful is image-based active learning for plant organ segmentation? Plant Phenomics. (2022). https://doi.org/10.3413/2022/9795275
Zhao, Z., et al.: Self-supervised assisted active learning for skin lesion segmentation, pp. 5043–5046. IEEE (2022). https://doi.org/10.1109/EMBC48229.2022.9871734
Jin, Q., Yuan, M., Qiao, Q., Song, Z.: One-shot active learning for image segmentation via contrastive learning and diversity-based sampling. Knowl.-Based Syst. 241, 108278 (2022). https://doi.org/10.1016/j.knosys.2022.108278
Radiuk, P.M.: Impact of training set batch size on the performance of convolutional neural networks for diverse datasets (2017). https://doi.org/10.1515/itms-2017-0003
Kandel, I., Castelli, M.: The effect of batch size on the generalizability of the convolutional neural networks on a histopathology dataset. ICT Express 6, 312–315 (2020). https://doi.org/10.1016/j.icte.2020.04.010
Zhang, Z., Liu, Q., Wang, Y.: Road extraction by deep residual u-net. IEEE Geosci. Remote Sens. Lett. 15, 749–753 (2018). https://doi.org/10.1109/LGRS.2018.2802944
Jha, D., et al.: Resunet++: An Advanced Architecture for Medical Image Segmentation, pp. 225–2255. IEEE (2019). https://doi.org/10.1109/ISM46123.2019.00049
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
AS conducted the experiments and drafted the manuscript, FA and OB participated in completing the analysis. All authors contributed to the research study. All authors reviewed the findings and approved the final manuscript version.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing financial or non-financial interests in the manuscript.
Consent for Publication
Mr. Abdelrahman I. Saad drafted the manuscript, Dr. Fahima A. Maghraby and Dr. Osama Badawy participated in all experiments. The final manuscript was read and approved by all of the authors.
Ethical Approval and Consent to Participate
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Saad, A.I., Maghraby, F.A. & Badawy, O. PolySeg Plus: Polyp Segmentation Using Deep Learning with Cost Effective Active Learning. Int J Comput Intell Syst 16, 148 (2023). https://doi.org/10.1007/s44196-023-00330-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44196-023-00330-6