Abstract
Purpose
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer. It is still unclear that the mechanisms by which long non-coding RNA (lncRNA) regulates tumorigenesis of TNBC. In this study, we explored the function and regulation of lncRNA in TNBC.
Methods
The differentially expressed and overlapped lncRNAs were obtained from two microarray datasets of breast cancer. The cancer genome atlas (TCGA) data was applied to validate the roles of top differentially expressed lncRNAs. The potential relationship among lncRNAs, miRNAs, and mRNAs and the effects of them on the TNBC tumorigenesis were further explored through multiple bioinformatic methods.
Results
Long intergenic non-protein coding RNA 1351 (LINC01351) was first discovered to play an oncogenic role in TNBC. The highly expressed LINC01351 was significantly related to aggressive subtypes, advanced stages and metastasis of breast cancer. The overexpressed LINC01351 was associated with adverse prognosis of patients with TNBC. LINC01351 was found to negatively regulate ELK4 which was involved in the transcriptional regulation in cancer. The high expression of ELK4 showed favorable prognosis of patients with basal-like 1 subtype and luminal androgen receptor subtype of TNBC.
Conclusion
The dysregulation of LINC01351 played an important role in triple-negative breast cancer. LINC01351 could be a poor prognostic marker and a potential target for patients with TNBC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Breast cancer is the most common malignant tumor and the second cause of cancer-related mortality in women [1]. Since 2015, breast cancer has become the top cancer of new cancer cases and deaths in China [2]. Triple-negative breast cancer (TNBC), which accounts for 15%-20% of breast cancer, is an aggressive subtype characterized by negative expression of estrogen receptor (ER), progesterone receptor (PgR) and ErbB-2/human epidermal growth factor receptor 2 (Her2) [3]. It has been well known that TNBC is associated with earlier metastasis, faster tumor proliferation, more drug resistance and poorer prognosis [4]. Except for poly (ADP-ribose) polymerase (PARP) inhibitors that have been used to treat patients with genomic instability and/or BRCA mutations, the therapies of TNBC are restricted to conventionally available chemotherapies [5]. In recent years, immunotherapy agents showed impressive result of clinical trials in the treatment of metastatic TNBC with positive expression of programmed cell death ligand 1 (PD-L1), however, it still need more validation to stratify the patient with immune infiltration [6]. It is urgently needed to uncover the mechanism of TNBC and to identify the novel prognostic target and therapeutic strategy for patients with TNBC.
Long non-coding RNAs (lncRNA) are a class of RNA transcripts with a length of > 200 nucleotides that do not encode proteins [7]. The function of lncRNA is involving in the flux of genetic information and diverse biological processes, such as splicing, stability of messenger RNA, epigenetic modulation, transcriptional, and post-translational modifications and cell proliferation and differentiation [7, 8]. Moreover, lncRNA is also reported to regulate tumorigenesis of multiple human cancers including breast cancer [9, 10]. However, the mechanism of lncRNA in TNBC has not been well investigated. In this study, we explored the function and regulation of lncRNA in TNBC to obtain the candidate prognostic marker and molecular target of therapy.
2 Materials and methods
2.1 Microarray data
Two lncRNA microarray datasets (GSE60689, TNBC vs. normal breast and GSE112848, breast cancer vs. normal breast) were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) [11, 12]. From GSE60689 dataset, the expression of lncRNAs was profiled and generated from the comparison of two stage III TNBC tissues and their paired adjacent noncancerous tissues by LncRNA Array 3.0 (ArrayStar). The pathological type of TNBC tissues was infiltrating ductal carcinoma. The ages of two female patients are 67 and 73, separately. The lncRNAs expression of GSE112848 were profiled and generated from three pairs of primary breast cancer tissues and adjacent noncancerous tissues. The pathological type of cancer tissues was ductal carcinoma in situ. The ages of three female patients are 43, 67 and 73, separately. Raw data was processed by SangerBox package (http://sangerbox.com/), GEO2R online software (https://www.ncbi.nlm.nih.gov/geo/geo2r/) and MultiExperiment Viewer (http://mev.tm4.org). The differentially expressed and overlapped lncRNAs with the criteria of |log fold change (FC)|> 3.0 and p value < 0.05 was screened out for further analysis.
2.2 Validation and target prediction of lncRNA
The top differentially expressed and overlapped lncRNAs were annotated and validated via UCSC (http://genome.ucsc.edu/), BLAST (https://blast.ncbi.nlm.nih.gov), and NONCODE (http://www.noncode.org/) online databases. Unannotated lncRNAs were removed from our data. Then the candidate lncRNAs were further detected through the Atlas of ncRNA in cancer (TANRIC) database (https://ibl.mdanderson.org/tanric/_design/basic/index.html). The target microRNAs (miRNAs) or mRNAs of candidate lncRNAs were predicted and analyzed by the TANRIC database. The potential interaction between miRNAs and their target mRNAs were explored by both miRTarBase (http://mirtarbase.mbc.nctu.edu.tw) and TargetScan (http://www.targetscan.org) online databases. Cytoscape software was used to visualize the regulatory network among lncRNAs, mRNAs and miRNAs. The functional enrichment of target mRNAs was analyzed by Metascape database (http://metascape.org).
2.3 Functional analysis of lncRNA
The correlation between the expression of lncRNA and the clinical characteristics including TNM stages, molecular subtypes, overall survival (OS) and relapse-free survival (RFS) were investigated with the data from The Cancer Genome Atlas (TCGA) Breast Cancer (BRCA) RNAseq, which was obtained from TANRIC database and UCSC Xena database (https://xenabrowser.net/) online databases. The survival analysis was performed by Kaplan–Meier Plotter (http://kmplot.com/analysis/) software. All the above databases were applied for in-depth analysis on the function and regulation of lncRNA.
2.4 Statistical analysis
The results were shown as mean ± standard deviation. The statistical significance of the microarray result was analyzed by fold change and the student's t-test. The threshold value we used to screen differentially expressed lncRNA is a fold change > 3.0 or < -3.0 (p < 0.05). The t-test was performed to analyze the difference between groups. Pearson correlation was applied to calculate different parameters in terms of correlation. The p value < 0.05 was considered as statistically significant. All analyses were conducted in SPSS Statistics 22.
3 Results
3.1 Dysregulated lncRNAs were found in triple-negative breast cancer
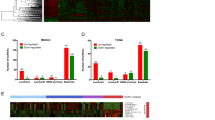
Two microarray datasets (GSE60689 and GSE112848) were analyzed according to the flow chart. As shown in Fig. 1, 3496 upregulated and 2677 downregulated lncRNAs were detected in breast cancer tissues comparing with normal breast tissues (BC groups). 723 upregulated and 753 downregulated lncRNAs were found in triple-negative breast cancer tissues comparing with normal breast tissues (TNBC groups). The results were shown as volcano plots according to the criteria of |log fold change (FC)|≥ 1.0 and p value ≤ 0.05 (Fig. 2A). In addition, 162 downregulated lncRNAs and 170 upregulated lncRNAs were investigated to be overlapped lncRNAs in both BC groups and TNBC groups. The overlapped lncRNAs were shown as the Venn diagram (Figs. 1 and 2B). Among them, the top differentially expressed lncRNAs including 4 downregulated lncRNAs and 17 upregulated lncRNAs were screened out and demonstrated as heatmaps with the criteria of |logFC|≥ 3.0 and p value ≤ 0.05 (Figs. 1 and 2C). The 21 lncRNAs were applied for further analysis. As shown in Fig. 1, after the annotation and verification of candidate 21 lncRNAs, three upregulated lncRNAs (LINC02303, LINC01351 and LINC01511) and two downregulated lncRNAs (CHL1-AS2 and LINC01612) were obtained from the strict verification via NONCODE and BLAST online software. Based on the literatures about the five dysregulated lncRNAs, the long intergenic non-protein coding RNA 1351 (LINC01351) was reported to be associated with the adverse prognosis of patient with colorectal cancer [13]. However, the effect of LINC01351 in breast cancer was not clear. Thus, LINC01351 was chosen for the following studies about the potential roles on breast cancer, especially TNBC.
The flow chart of GEO datasets analysis. The expression data of lncRNA from two Gene Expression Omnibus (GEO) datasets including GSE60689 and GSE112848 was processed and analyzed according to flow diagram. TNBC, triple-negative breast cancer; BC, breast cancer; lncRNA, long non-coding RNA; FC, fold change
Dysregulated lncRNAs were found in triple-negative breast cancer. A The volcano plots of two lncRNA microarray datasets showed the differential expression of lncRNAs in breast cancer (BC group, breast cancer vs. normal) and triple-negative breast cancer (TNBC group, TNBC vs. normal). In BC group, 3496 lncRNAs were discovered to be upregulated, while 2677 lncRNAs were downregulated. In TNBC group, 723 lncRNAs were identified to be upregulated, while 753 lncRNAs were downregulated. The red points represented upregulated lncRNAs, while the green points represented downregulated lncRNAs. The black points represented lncRNAs with no significant difference. Both P-value < 0.05 and |log fold change (FC)|> 1 was considered to be significantly different. BC, breast cancer; TNBC, triple-negative breast cancer; Down, downregulated lncRNAs; Up, upregulated lncRNAs. B The Venn diagram showed the overlaps of differentially expressed lncRNAs from the BC group (BC vs. normal) and TNBC group (TNBC vs. normal). 162 lncRNAs were found to be overlapped and downregulated in both BC and TNBC groups, while 170 lncRNAs were detected to be upregulated. BC, breast cancer; TNBC, triple-negative breast cancer; Down, downregulated lncRNAs; Up, upregulated lncRNAs. C The cluster heatmaps of two lncRNA microarray datasets showed the differentially expressed lncRNAs in both BC and TNBC groups. The red blocks represented upregulated lncRNAs, while the green blocks represented downregulated lncRNAs. 17 lncRNAs were found to be upregulated, while 4 lncRNAs were detected to be downregulated in both BC and TNBC groups with the criteria of both P-value < 0.05 and |log fold change (FC)|> 3. BC, breast cancer; TNBC, triple-negative breast cancer
3.2 LINC01351 was an oncogenic indicator in breast cancer
To date, a large number of lncRNAs have been discovered through functional genomic studies, however, the potential role of LINC01351 in breast cancer has not been well investigated yet. The function and regulation of LINC01351 were explored in the study. The expression of LINC01351 was tested by using the data from GEO datasets (GSE60689 and GSE112848). All the data was processed to be two groups including BC groups (BC vs. normal breast) and TNBC groups (TNBC vs. normal breast). As shown in Fig. 3A-a/b, LINC01351 was highly expressed in both BC tissues or TNBC tissues compared with normal breast tissues (BC vs. normal, p = 0.007; TNBC vs. normal, p = 0.034). To further test the regulation of expression, the data from GDC TCGA breast cancer (BRCA) database was induced to be analyzed. 49 cases of normal breast samples, 595 cases of breast cancer samples, and 8 cases of TNBC samples were included in the analysis. Similarly, LINC01351 was found to be overexpressed in TNBC and breast cancer tissues comparing to normal breast tissues (Fig. 3A-c, all p < 0.001).
LINC01351 was an oncogenic marker in breast cancer. A High-expressed LINC01351 was found in BC tissues or TNBC tissues compared with normal breast tissues. a LINC01351 was higher expressed in BC tissues than normal breast tissues by using data from GSE112848 datasets (p = 0.007). b LINC01351 was higher expressed in TNBC tissues than normal breast tissues by using data from GSE60689 datasets (p = 0.034). c LINC01351 was found to be higher expressed in BC tissues or TNBC tissues than normal breast tissues by using the GDC TCGA BRCA data (p < 0.001). B Increased expression of LINC01351 was associated with aggressive subtypes of breast cancer. Higher expression of LINC01351 was discovered in ER-negative, PR-negative or Her2-positive breast cancer tissues than ER-positive, PR-positive or Her2-negative breast cancer tissues (p < 0.001). C The expression of LINC01351 was higher in triple-negative breast cancer tissues than in luminal-subtype breast cancer tissues (p < 0.05). D For the TNM stages of breast cancer, the expression of LINC01351 was progressively increasing from stage I/II to stage III/IV (p = 0.007). E Higher expression of LINC01351 was detected in metastatic breast cancer tissues than in primary breast cancer tissues by using the GDC TCGA BRCA data (p = 0.003)
In breast cancer, hormone-receptor negative (ER and PR negative) and Her2 positive are considered to be poor prognostic markers. So, the correlation between the expression of LINC01351 and molecular pathological features of breast cancer were further detected via TCGA and TANRIC database. As shown in Fig. 3B, increased expression of LINC01351 was discovered in ER-negative, PR-negative and Her2-positive breast cancer tissues (ER-negative vs. ER-positive, p = 3.21e-4; PR-negative vs. PR-positive, p = 9.30e-8; Her2-positive vs. Her2-negative, p = 3.83e-8), which suggested that LINC01351 contributed to the malignant phenotypes of breast cancer. In the GDC TCGA BRCA data, 18 cases of luminal A, 24 cases of luminal B, 10 cases of Her2-enriched and 15 cases of TNBC samples were induced into the analysis. Similarly, the expression of LINC01351 was significantly higher in Her2-enriched or triple-negative breast cancer tissues than in luminal subtypes of breast cancer tissues (TNBC vs. luminal A, p = 0.012; TNBC vs. luminal B, p = 0.045; Fig. 3C). There was no significant difference in the expression of LINC01351 between Her2-enriched and triple-negative breast cancer (TNBC vs. Her2-positive, p = 0.073).
Moreover, the expression of LINC01351 was progressively increasing with the TNM stages of breast cancer (p = 0.007, Fig. 3D). Generally, TNBC is an aggressive subtype of breast cancer with early metastasis. Then the expression of LINC01351 was further detected in both metastatic tumor samples and primary tumor samples by exploiting the GDC TCGA BRCA data. The higher expressed LINC01351 was found in the metastatic breast cancer tissues than in the primary breast cancer tissues (Fig. 3E, p = 0.003).
All these results indicated that highly expressed LINC01351 contributed to the aggressive subtypes, advanced TNM stages, and metastasis of breast cancer. It was the first time to report that LINC01351 played as an oncogenic and poor molecular pathological indicator of breast cancer.
3.3 LINC01351 was a poor prognostic marker in triple-negative breast cancer
To understand the potential role of LINC01351 on the survival of patients with breast cancer, the survival data was further analyzed via the TCGA and TANRIC database. The primary breast cancer tissues were divided equally into two groups based on the expression of LINC01351 (low-expression and high-expression, n = 372, separately). In the study, all the duration of following-up was 10 years. As demonstrated in Fig. 4A, the result of survival analysis showed that LINC01351 played no effect on the overall survival (OS) of breast cancer (p = 0.3265). For the relapse-free survival (RFS), 588 cases of breast cancer with survival data were introduced into the analysis. Low-expressed group (n = 294) and high-expressed group (n = 294) were also divided by the expression of LINC01351. LINC01351 showed no effect on the RFS of breast cancer (p = 0.06, Fig. 4B).
LINC01351 was a poor prognostic indicator in triple-negative breast cancer. A LINC01351 played no effect on the overall survival of breast cancer after 10-year-following up (p = 0.3265). B LINC01351 showed no effect on the relapse-free survival of breast cancer after 10-year-following up (p = 0.06). C Higher expressed LINC01351 was associated with poor overall survival of patients with TNBC after 10-year-following up (p = 0.047). D LINC01351 was significantly related to unfavorable relapse-free survival of patients with TNBC after 10-year-following up (p = 0.029)
Then for the analysis about TNBC, the higher expression of LINC01351 was found to be significantly related with poorer prognosis of patients. 83 cases of TNBC were divided into low-expressed group (n = 42) and high-expressed group (n = 41). Higher expressed LINC01351 was associated with poor OS of patients with TNBC (p = 0.047, Fig. 4C). Similarly, LINC01351 was significantly related to poor RFS of patients with TNBC from the analysis about 30 cases of low-expressed TNBC and 29 cases of high-expressed TNBC (p = 0.029, Fig. 4D).
All the above data indicated that highly expressed LINC01351 was associated with poor prognosis of patients with TNBC. It was suggested that the role of LINC01351 on survival of patients was specific for TNBC but not for whole types of breast cancer. LINC01351 was an unfavorable prognostic marker of TNBC.
3.4 Integrative analysis of lncRNA, mRNA and microRNA signature
Long non-coding RNA presents interaction domains for DNA, mRNA, microRNA (miRNA, miR), and protein, depending on both sequence and secondary structure [8]. Emerging evidence indicates that lncRNA could be ceRNA for miRNAs in cancer [14]. Thus, we explored the potential regulation of LINC01351 to mRNAs and miRNAs. The network of regulation was inquired and further visualized by Cytoscape software. Among them, 6 miRNAs (miR-504, miR-548n, miR-567, miR-618, miR-718, and miR-1280) and 4 mRNAs (ELK4, ALPI, SCEMOL and RAD23B) were identified to be targets of LINC01351. Based on the analysis from TANRIC database, LINC01351 downregulated ELK4, SCEMOL and RAD23B and upregulated ALPI (all p < 0.001). For the miRNAs, miR-504 was downregulated by LINC01351, while miR-548n, miR-567, miR-618, miR-718, and miR-1280 were upregulated by LINC01351 (all p < 0.01). The positive and negative regulation between LINC01351 and target mRNAs or miRNAs were all described in Fig. 5.
The regulatory network of lncRNA, mRNAs and miRNAs. For mRNAs, LINC01351 downregulated ELK4, SCEMOL and RAD23B and upregulated ALPI (all p < 0.001). For miRNAs, LINC01351 downregulated miR-504 and upregulated miR-548n, miR-567, miR-618, miR-718, and miR-1280 were by LINC01351 (all p < 0.01). All the target mRNAs were predicted to be downregulated by their miRNAs. Red, LINC01351; purple, downregulated mRNA; yellow, upregulated mRNA; grey, downregulated miRNAs; green, upregulated miRNAs and blue, the target mRNAs of miRNAs. MiRNA or miR, microRNA
It is well known that miRNAs are small non-coding RNAs that usually inhibit gene expression through partially complementary elements in the 3′ UTR of their target mRNAs [15]. Furthermore, the downstream target genes of six miRNAs (miR-504, miR-548n, miR-567, miR-618, miR-718, and miR-1280) were obtained from TargetScan and miRTarBase database. All the target mRNAs were predicted to be downregulated by their miRNAs. Then the overlapped target mRNAs of both TargetScan and miRTarBase databases were also shown by Cytoscape software in Fig. 5. The above result demonstrated that LINC01351 was involved in the complex regulation network of mRNAs and miRNAs.
3.5 ELK4 was down-regulated by LINC01351 and associated with transcriptional regulation in cancer
To further understand the effect of LINC01351 during carcinogenesis, functional enrichment and GO annotations of both overlapped genes and target genes of LINC01351 were analyzed by Metascape database. It was found that gene signature was enriched in the transcriptional misregulation in cancer (p < 0.001, Fig. 6A). In the enriched functional group ETS transcription factor ELK4, also known as SAP1, was chosen for the following analysis, because it was identified as the direct target negatively regulated by LINC01351 (p = 0.00034). ETS-like transcription factor 4 (ELK4) is a member of the ETS family of transcription factors and of the ternary complex factor (TCF) subfamily [16]. The androgen respondent ELK4 was reported to be correlated with the oncogene MYC [17]. However, the exact role of ELK4 in TNBC was not well understood yet.
LINC01351/ELK4 axis predicted prognosis of patients and sensitivity of therapy in triple-negative breast cancer. A The functional analysis showed that gene signature of candidate mRNAs was enriched in the transcriptional misregulation of cancer. B Representative result of ELK4 expression. ELK4 was high-expressed in normal breast tissue and weak-expressed in breast cancer tissue by immunohistochemistry. C The expression of ELK4 was negatively correlated with the expression of LINC01351 in breast cancer (p = 0.019). D The high expression of ELK4 was associated with the favorable overall survival of patients with TNBC (p = 0.0035). Moreover, higher expression of ELK4 was significantly related to better overall survival in basal-like 1 (BL1) and luminal androgen receptor (LAR) subtypes of TNBC after 10-year-following up (all p < 0.05)
To clarify the effect of ELK4 and the inhibition role regulated by LINC01351, the expression of ELK4 was tested via immunohistochemistry (IHC) in breast cancer tissues compared with normal breast tissues. As shown in Fig. 6B, the results of IHC showed that ELK4 was high-expressed in normal breast tissue and weak-expressed in breast cancer tissue, which was exactly opposite to the expression of LINC01351 in breast cancer compared with normal breast tissue. Moreover, the correlation between the expression of ELK4 and LINC01351 was investigated by analyzing the TCGA and TANRIC database. As shown in Fig. 6C, the expression of ELK4 showed negative correlation with the expression of LINC01351 (p = 0.019). All the results demonstrated that LINC01351 down-regulated the expression of ELK4 in breast cancer.
3.6 LINC01351/ELK4 axis predicted prognosis and sensitivity of therapy in triple-negative breast cancer
In the study, to explore the regulation of LINC01351/ELK4 axis on TNBC, the survival analysis was performed by using Kaplan–Meier Plotter database. The high expression of ELK4 was associated with the favorable prognosis of patients with TNBC (p = 0.0035, Fig. 6D). TNBC is defined by lack of expression of ER, PR and Her2 and is still a big challenge for anticancer therapy. To identify the best treatment strategy, 6-subtype-classification of TNBC was reported by Lehmann in 2011 [18]. For each subtype, available therapy was predicted and recommended in the study. Based on the classification, TNBC was classified as basal-like 1 (BL1), basal-like 2 (BL2), immunomodulatory (IM), mesenchymal (M), mesenchymal stem-like (MSL), and luminal androgen receptor (LAR) subtype. Furthermore, the relationship between the expression of ELK4 and different subtypes of TNBC (BL1, BL2, IM, M, MSL and LAR) was detected in the following analysis. The survival results demonstrated that higher expression of ELK4 was significantly related to better overall survival in BL1 and LAR subtypes of TNBC after 10-year-following up (all p < 0.05, Fig. 6D). The survival data was exactly opposite to the survival data of LINC01351 in TNBC, which suggested that LINC01351 negatively regulated the expression of ELK4 and affected its roles on the overall survival of patients with BL1 and LAR subtypes of TNBC. Besides, LINC01351/ELK4 axis could be a potential prognostic marker of TNBC.
Based on the subtypes of Lehmann classification, BL1 and BL2 subtypes preferentially respond to the therapy regulating the cell cycle and DNA damage. The patient with BL1 subtype of TNBC could receive more benefits from chemotherapy. IM subtype is associated with immunotherapy. M and MSL subtypes are related to epithelial-mesenchymal transition, growth factor pathways and PI3K/mTOR pathway. The LAR subtype is characterized by androgen receptor (AR) signaling and sensitive to AR antagonist [18]. The above results showed that the higher expression of ELK4 was significantly related to better overall survival in BL1 and LAR subtypes of TNBC. The BL1 or LAR subtype of TNBC with low-expressed LINC01351 and high-expressed ELK4 was considered to be more sensitive to the treatment of chemotherapy (e.g., cisplatin) or AR antagonist drugs (e.g., bicalutamide), separately. It could be an easy approach to test the expression of LINC01351/ELK4 axis to predict the prognosis and sensitivity of anticancer therapy.
4 Discussion
Long non-coding RNAs (lncRNA) are non-coding transcripts > 200 nucleotides long that have been found to act as master regulators for gene expression and play a critical role in various biological functions and diseases including human cancers. Several studies have identified that lncRNA with predictive properties of cancer progression or diagnostic power in breast cancer [19]. In eukaryotic cells, long intergenic non-coding RNAs (lincRNAs) are the largest class of lncRNAs. The transcripts of lincRNAs do not overlap protein-coding sequences [20]. Increasing evidence suggests that lincRNAs play effects on the maintenance of cell integrity, regulation of gene function, and involvement in development and disease. Marked upregulation of lincRNAs such as Linc-ROR was observed in breast cancer [21, 22]. However, the function and regulation of long intergenic non-protein coding RNA 1351 (LINC01351) in breast cancer has not been reported yet.
Here, we found that LINC01351 was high-expressed in triple-negative breast cancer tissues compared with normal breast tissues or luminal-subtype breast cancer tissues. The overexpressed LINC01351 was positively correlated with aggressive subtypes, advanced TNM stages, and metastasis of breast cancer. Additionally, the upregulated expression of LINC01351 predicted unfavorable overall survival and relapse-free survival of patients with TNBC. It was the first time that LINC01351 was discovered to be a poor prognostic indicator and an oncogenic marker in triple-negative breast cancer.
In our study, LINC01351 was found to down-regulate the expression of ELK4. ELK4 is a member of the ternary complex factor subfamily of E twenty-six domain transcription genes [16]. ELK4 was reported to be correlated with the oncogene MYC in breast cancer [17]. In colorectal cancer, ELK4 cooperated with BRD2 to regulate the transcription of LAMB [23]. ELK4 was involved in the regulation of the chimeric fusion RNA, SLC45A3-ELK4, and played effects on cell proliferation in androgen-dependent prostate cancer cells [24]. However, the exact role of ELK4 in TNBC was not well understood. In our studies, ELK4 and LINC01351 showed the inverse impact on overall survival of patient with TNBC. High-expression ELK4 was associated with favorable prognosis of patients. Furthermore, higher expression of ELK4 was significantly related to better overall survival in BL1 and LAR subtypes of TNBC after 10-year-following up. All the above finding showed that LINC01351 negatively modulated ELK4 in triple-negative breast cancer. The upregulation of LINC01351 was a poor prognostic marker for patient with triple-negative breast cancer. In addition, the BL1 or LAR subtype of TNBC with low-expressed LINC01351 and high-expressed ELK4 could be more sensitive to the treatment of chemotherapy or AR antagonist drugs. The potential relationship is worth to explore in future studies.
5 Conclusion
In our study, it revealed that highly expressed LINC01351 was significantly related to aggressive subtypes, advanced stages and metastasis of breast cancer. LINC01351 was an oncogenic indicator and a poor prognostic marker in TNBC by negatively regulating ELK4. The high expression of ELK4 showed favorable prognosis of patients with BL1 and LAR subtype of TNBC. A novel regulatory axis was revealed, which may offer new insight into the prognostic evaluation and treatment strategies for patients with triple-negative breast cancer.
Availability of data and materials
All data generated or analyzed during this study are all included in this manuscript.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52.
Arnold KM, Pohlig RT, Sims-Mourtada J. Co-activation of Hedgehog and Wnt signaling pathways is associated with poor outcomes in triple negative breast cancer. Oncol Lett. 2017;14(5):5285–92.
Papadimitriou M, Mountzios G, Papadimitriou CA. The role of PARP inhibition in triple-negative breast cancer: Unraveling the wide spectrum of synthetic lethality. Cancer Treat Rev. 2018;67:34–44.
Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Diéras V, Hegg R, Im SA, Shaw Wright G, Henschel V, Molinero L, Chui SY, Funke R, Husain A, Winer EP, Loi S, Emens LA. IMpassion130 trial investigators atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N Engl J Med. 2018;379(22):2108–21.
Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62.
Fernandes JCR, Acuna SM, Aoki JI, Floeter-Winter LM, Muxel SM. Long Non-Coding RNAs in the Regulation of Gene Expression: Physiology and Disease. Noncoding RNA. 2019;5(1):17.
Zhang R, Xia LQ, Lu WW, Zhang J, Zhu JS. LncRNAs and cancer. Oncol Lett. 2016;12(2):1233–9.
Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, Wang C, Hawke DH, Wang S, Zhang Y, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18(2):213–24.
Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, Park PK, Qin L, Wei Y, Hawke DH, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159(5):1110–25.
Krishnan P, Ghosh S, Wang B, Li D, Narasimhan A, Berendt R, Graham K, Mackey JR, Kovalchuk O, Damaraju S. Next generation sequencing profiling identifies miR-574-3p and miR-660-5p as potential novel prognostic markers for breast cancer. BMC Genomics. 2015;16:735.
Qin F, Xu H, Wei G, Ji Y, Yu J, Hu C, Yuan C, Ma Y, Qian J, Li L, Huo J. A Prognostic Model Based on the Immune-Related lncRNAs in Colorectal Cancer. Front Genet. 2021;12:658736.
Dong J, Teng F, Guo W, Yang J, Ding G, Fu Z. lncRNA SNHG8 Promotes the Tumorigenesis and Metastasis by Sponging miR-149-5p and Predicts Tumor Recurrence in Hepatocellular Carcinoma. Cell Physiol Biochem. 2018;51(5):2262–74.
Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci U S A. 2006;103(8):2746–51.
Zheng L, Xu H, Di Y, Chen L, Liu J, Kang L, Gao L. ELK4 promotes the development of gastric cancer by inducing M2 polarization of macrophages through regulation of the KDM5A-PJA2-KSR1 axis. J Transl Med. 2021;19(1):342.
Mesquita B, Lopes P, Rodrigues A, Pereira D, Afonso M, Leal C, Henrique R, Lind GE, Jerónimo C, Lothe RA, Teixeira MR. Frequent copy number gains at 1q21 and 1q32 are associated with overexpression of the ETS transcription factors ETV3 and ELF3 in breast cancer irrespective of molecular subtypes. Breast Cancer Res Treat. 2013;138(1):37–45.
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–67.
Su X, Malouf GG, Chen Y, Zhang J, Yao H, Valero V, Weinstein JN, Spano JP, Meric-Bernstam F, Khayat D, et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget. 2014;5(20):9864–76.
Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–5.
Loewen G, Zhuo Y, Zhuang Y, Jayawickramarajah J, Shan B. lincRNA HOTAIR as a novel promoter of cancer progression. J Can Res Updates. 2014;3(3):134–40.
Eades G, Wolfson B, Zhang Y, Li Q, Yao Y, Zhou Q. lincRNA-RoR and miR-145 regulate invasion in triple-negative breast cancer via targeting ARF6. Mol Cancer Res. 2015;13(2):330–8.
Zhu Z, Song J, Guo Y, Huang Z, Chen X, Dang X, Huang Y, Wang Y, Ou W, Yang Y, et al. LAMB3 promotes tumour progression through the AKT-FOXO3/4 axis and is transcriptionally regulated by the BRD2/acetylated ELK4 complex in colorectal cancer. Oncogene. 2020;39(24):4666–80.
Qin F, Zhang Y, Liu J, Li H. SLC45A3-ELK4 functions as a long non-coding chimeric RNA. Cancer Lett. 2017;404:53–61.
Code availability
The microarray datasets (GSE60689 and GSE112848) analyzed in this paper were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/).
Funding
This work was supported by the following grants: Foundation of Young Innovative Talents (Tianjin Medical University Cancer Institute and Hospital, 2018–1-31); National Science Foundation of China (NSFC No. 81401950), China Scholarship Council (File No. 201706940019), Tianjin Key Medical Discipline (Specialty) Construction Project, and Tianjin Medical University Cancer Hospital "14th Five-Year" Peak Discipline Support Program Project.
Author information
Authors and Affiliations
Contributions
Y.J., Ys.S. and Z.T. conceived the research design. Y.J. and Ys.S. were the main contributors of manuscript text and figures. Y.J., Ys.J., Yh.S., W.Z. and X.W. performed all the data mining and analysis. J.M. revised and polished the language. All authors reviewed the manuscript, provided edits and suggestions, and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All data in this manuscript was collected under the guidelines approved by the institutional review board of Tianjin Medical University Cancer Institute and Hospital and complying with the current laws in China. No sample from human or animal was directly used in this study.
Consent for publication
All authors agree with the submission in its present (and subsequent) forms.
Competing interests
The authors report no conflict of interest in this work.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jia, Y., Jia, Y., Shi, Y. et al. Integrative analysis of LncRNA-mRNA signature reveals a functional LincRNA in triple-negative breast cancer. Holist Integ Oncol 1, 9 (2022). https://doi.org/10.1007/s44178-022-00009-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44178-022-00009-6