Abstract
The type VI secretion system (T6SS) is a powerful bacterial molecular weapon that can inject effector proteins into prokaryotic or eukaryotic cells, thereby participating in the competition between bacteria and improving bacterial environmental adaptability. Although most current studies of the T6SS have focused on animal bacteria, this system is also significant for the adaptation of plant-associated bacteria. This paper briefly introduces the structure and biological functions of the T6SS. We summarize the role of plant-associated bacterial T6SS in adaptability to host plants and the external environment, including resistance to biotic stresses such as host defenses and competition from other bacteria. We review the role of the T6SS in response to abiotic factors such as acid stress, oxidation stress, and osmotic stress. This review provides an important reference for exploring the functions of the T6SS in plant-associated bacteria. In addition, characterizing these anti-stress functions of the T6SS may provide new pathways toward eliminating plant pathogens and controlling agricultural losses.
Similar content being viewed by others
Introduction
The relationships among bacteria, their hosts, and other microbes in the environment (Stubbendieck et al. 2016) determine the conditions for bacterial adaptation to the environment determine the conditions for bacterial adaptation to the environment. Bacteria must protect themselves from adverse environmental conditions and host immune system attacks (Tang et al. 2022). Bacteria have evolved diverse survival mechanisms in response to selection pressures, including the use of protein secretion systems to combat environmental stresses (Kempnich and Sison-Mangus. 2020). The bacterial type VI secretion system (T6SS) is a contact-dependent injection mechanism that usually targets bacterial competitors (Mougous et al. 2006; Pukatzki et al. 2006; Le Goff et al. 2022) and eukaryotic hosts (Hood et al. 2010; Liu et al. 2023).
The T6SS system was originally found in plant-associated bacteria, where it has many important functions, including helping the bacteria cope with stresses from other bacteria or the plant hosts (Bladergroen et al. 2003; Gallegos-Monterrosa and Coulthurst. 2021; Wang et al. 2021b). In this paper, we review the structural assembly of T6SS and the involvement of T6SS in the stress resistance of plant-associated bacteria. In addition, the role of T6SS in the resistance of plant-associated bacteria to plant hosts is discussed.
T6SS
The T6SS is a syringe structure similar to a contractile phage tail that usually consists of 13 conserved components, named TssA to TssM, forming a structure that is capable of secreting proteins (effectors) into the target cell or the extracellular environment (Fig. 1) (Cherrak et al. 2019). The structure of T6SS consists of the transmembrane complex, the baseplate complex, and the tail-tube complex (Ho et al. 2014; Basler 2015; Nguyen et al. 2017; Taylor et al. 2018). The transmembrane complex is formed by several transmembrane proteins. Most bacterial T6SS gene clusters encode three membrane proteins: TssJ, TssL, and TssM. TssL and TssM are anchored in the inner membrane, while TssJ, as an outer membrane-associated lipoprotein, exists in the periplasm (Aschtgen et al. 2008; Felisberto-Rodrigues et al. 2011). The baseplate complex is wedge-shaped and is composed of TssK, TssF, TssG, and TssE. There is a close relationship between the baseplate complex and other structures, and its function is to anchor the tail-tube complex to the transmembrane complex to initiate T6SS assembly (Zoued et al. 2016; Cherrak et al. 2018). The tail-tube complex is built on the baseplate complex and can be assembled in a short time, forming a tubular structure in the cytoplasm that includes the Hcp (TssD) inner tube, the TssB-TssC outer sheath, and the VgrG (TssI)-PAAR tip complex located at the top of the sheath (Shneider et al. 2013; Ho et al. 2014). The assembly of the T6SS is thought to begin with the assembly of the transmembrane complex, followed by the recruitment of the baseplate complex and the extension of the sheath (Wang et al. 2019). First, the outer membrane lipoprotein TssJ recruits the inner membrane proteins TssM and TssL to form a transmembrane complex (Cherrak et al. 2018). Next, the inner tube and sheath of the tail-tube complex are coordinated for assembly, where the Hcp protein and TssBC are recruited to the baseplate complex and are stacked continuously, while the TssBC sheath is rapidly polymerized to establish a tubular structure about one micron long (Basler et al. 2012). The contraction of the sheath pushes the inner tube of Hcp, the VgrG-PAAR complex, and the carried effector protein to the target cell, thereby completing the transport of the effector protein (Ruhe et al. 2020). After the delivery of the effector protein, ClpV (TssH) AAA+ ATPase binds to the contracted TssBC outer sheath for disassembly into subunits that can be recycled for the next T6SS assembly (Pietrosiuk et al. 2011; Kapitein et al. 2013).
The T6SS system was first discovered and studied in the plant-associated bacterium Rhizobium leguminosarum that affects pea nodulation and nitrogen fixation (Bladergroen et al. 2003). However, with the deepening of research, the T6SS has been demonstrated to have diverse functions (Lin et al. 2021), including anti-bacterial (Trunk et al. 2018) and anti-host activities (Coulthurst. 2019), mediating the uptake of metal ions (Lin et al. 2017; Han et al. 2019), resistance to environmental stress (Si et al. 2017a; Wan et al. 2017), inhibition of bacterial infection in the host (Bendor et al. 2015), and regulating the formation of bacterial biofilms (Lin et al. 2015; Chen et al. 2020). At present, the literature on T6SS primarily focuses on animal bacteria, while there are fewer studies on the function of plant-associated bacterial T6SS (Wang et al. 2023).
T6SS confers resistance to biotic stresses
The T6SS exists in about 25% of gram-negative bacteria (Boyer et al. 2009; Bock et al. 2017), including a large number of plant-associated bacteria (Bernal et al. 2018). Plant-associated bacteria in the environment are a diverse group that includes plant-beneficial bacteria and plant pathogens (Cui et al. 2015). The T6SS functions in the resistance of plant-related bacteria to biotic stresses such as plant pathogens and adverse environmental conditions (Fig. 2; Table 1) (Turner et al. 2013; El-Saadony et al. 2022).
A model of T6SS involved in the biotic stress resistance of plant-associated bacteria. The T6SS participates in the competition with plant pathogenic bacteria by secreting effector proteins. The T6SS affects the virulence of plant pathogens by regulating the inter-bacterial competition, biofilm formation, and environmental adaptability. The T6SS can regulate the colonization of plant pathogens and affect the symbiosis of Rhizobium and host by affecting nodulation
Competition with plant pathogens
The T6SS plays an important role in the competition among bacteria (Santoriello and Pukatzki. 2023; Zhang et al. 2023). Most plant-associated bacteria that possess the T6SS have strong viability when they coexist with other bacteria (Bernal et al. 2018; Ripcke et al. 2023). The study of plant-associated bacterial T6SS shows that this competitive advantage is achieved by injecting toxic effector proteins into rival bacteria via the T6SS to kill or inhibit their growth (Whitney et al. 2013; Tang et al. 2018).
Agrobacterium tumefaciens, a soil bacterial species that triggers plant tumorigenesis (Huang et al. 2021), employs the T6SS to gain a competitive advantage. In an experiment on intra-plant infection, A. tumefaciens secreted the effector Tde that possesses anti-bacterial DNA enzyme activity via the T6SS, giving the bacteria an advantage in the competition between homologous cells and Pseudomonas aeruginosa. At the same time, the bactericidal effect of this effector protein can be counteracted by the cognate immunity protein Tdi in A. tumefaciens (Ma et al. 2014). The conservation of toxin effector protein–immune protein (EI) pairs in T6SS-dependent bacteria highlights the use of the T6SS as a widespread antibacterial weapon that is beneficial for niche colonization (Yang et al. 2018). Moreover, in addition to T6SS-related EI pairs that can affect the competitiveness of A. tumefaciens, recent studies have shown that genetic characteristics also have a significant impact on the antagonistic interactions between bacteria, thus opening new avenues for the study of the ecological role of competition among bacteria (Wu et al. 2019). In addition to plant pathogenic bacteria, a T6SS-mediated competitive advantage also exists in some non-pathogenic plant bacteria such as Pseudomonas putida (Durán et al. 2021). For example, the Tke2 protein in P. putida has been confirmed to be an effector protein secreted by the T6SS that belongs to the HNH nuclease superfamily and exhibits nuclease activity. This protein is dependent on K1-T6SS secretion and has an inhibitory effect on the growth of Escherichia coli when heterologously expressed in E. coli (Bernal et al. 2017). P. putida is thought to be able to outcompete plant pathogenic bacteria such as Xanthomonas campestris by the action of the T6SS. Although this phenomenon has been observed both in vitro and in plants, whether this depends on the effector proteins identified above remains to be verified (Bernal et al. 2017). As a plant-beneficial bacterium, Rhizobium can produce nutrients to promote plant growth and increase plant yield (Yang et al. 2022a). Recent studies have found that the T6SS also plays a role in the intracellular competition of Rhizobium and the competition between Rhizobium and other plant pathogens. A recent study identified a T6SS-dependent effector, Re78, and an immunity protein, Re79, in Rhizobium etli Mim1. When wild strains were co-cultured with re78/re79 mutants, the mutants were less competitive, indicating that Re78/Re79 were related to bacterial competition in Rhizobium etli Mim1 (De Sousa et al. 2023). There are two sets of T6SSs (T6SS-1 and T6SS-3) in another rhizobial strain, Paraburkholderia sabiae. In a competition experiment with several common plant pathogens (Pseudomonas syringae DC3000, Pectobacterium carotovorum LMG2404, and Ralstonia solanacearum DSM9544), the mutants in which the core gene tssC of T6SS-1 was mutated showed lower competitiveness compared with the wild-type strain (Hug et al. 2023), indicating that the T6SS played an important role in the competition between P. sabiae and other plant pathogens.
In conclusion, many plant-associated bacteria contain the T6SS, and they employ the T6SS to secrete toxic effector proteins to compete with other bacteria, allowing them to gain sufficient growth advantages in the complex bacterial environment. Although the effector proteins involved in interbacterial competition have not been identified in some bacteria, phenotypic analysis has shown that the T6SS confers advantages in competition (Boak et al. 2022).
Resistance to plant pathogens and plant pests
Crop diseases are long-term problems that cause serious economic losses and affect the sustainable supply of food (Savary and Willocquet. 2020). While chemical pesticides can prevent crop diseases, the excessive use of pesticides is harmful to both human and environmental health (Nanjani et al. 2022). Microbial agents have been recognized as feasible and environmentally friendly pesticide alternatives (Zhang et al. 2016). Recent studies have found that many plant-beneficial bacteria, as biocontrol agents, possess the T6SS (Marchi et al. 2013), and most of these beneficial plant bacteria belong to the genus Pseudomonas. These bacteria can inhibit plant diseases and are considered an important source of biocontrol agents (Abo-Elyousr et al. 2021; Al-Karablieh et al. 2022; Gu et al. 2022).
There is a set of T6SS in Pseudomonas fluorescens strain MFE01 that exerts antibacterial activity against many common rhizosphere bacteria (Decoin et al. 2014). MFE01 can inhibit Pectobacterium atrosepticums and resist potato tuber soft rot, while mutant T6SS strains lose the inhibitory effect on P. atrosepticums (Decoin et al. 2014). A recent study found that this inhibition was due to the fact that T6SS in MFE01 can secrete an effector protein Tae3Pf with amidase activity that has antibacterial activity and can thus protect potato tubers from P. atrosepticums infection (Bourigault et al. 2023). Therefore, T6SS in P. fluorescens MFE01 can inhibit the plant pathogen P. atrosepticums and indirectly protect potato tubers from plant diseases. In addition, P. putida is a saprophytic soil bacterium that can colonize the roots of crop plants; it is also one of the most characteristic biocontrol plant growth-promoting bacteria (Abo-Elyousr et al. 2021). There are three kinds of T6SS (K1-T6SS, K2-T6SS, and K3-T6SS) in P. putida KT2440, of which K1-T6SS is primarily used against other bacteria, including many plant pathogens (Bernal et al. 2017). P. putida KT2440 was used with X. campestris to infect Nicotiana benthamiana leaves, and green fluorescent protein-labeled X. campestris was used for in-situ colonization monitoring. Compared with the wild-type KT2440 strain, the rate of leaf necrosis and the number of X. campestris were increased when the mutant T6SS strain and green fluorescent protein-labeled X. campestris co-infected N. benthamiana leaves (Bernal et al. 2017). These results suggest that the protective effect of P. putida in plant leaves is due to the decreased survival rate of X. campestris in leaves, and this protection depends on the T6SS. The genome of Azospirillum brasilense contains two complete sets of genes encoding T6SS, and studies have shown that the T6SS of A. brasilense exhibits antagonistic activity in vitro against many plant pathogens, including Agrobacterium, Pectobacterium, Dickeya, and Ralstonia species; this allows the T6SS of A. brasilense to provide bio-control protection against microalgae and plants against bacterial pathogens (Cassan et al. 2021).
In addition to pathogenic Pseudomonas, other Pseudomonas bacteria also employ the T6SS to resist plant pests and eukaryote bacterivores, thereby demonstrating effective biological control. The T6SS in Pseudomonas protegens strain CHA0 functions in insect invasion and pathogenicity. Using larvae of the cabbage butterfly Pieris brassicaea as a plant-feeding insect model, the T6SS was demonstrated to be able to kill insects following oral infection. This killing effect was achieved by the T6SS and its secreted effector proteins through promoting the colonization of bacteria in the insect gut and the resulting competition with commensal gut bacteria (Vacheron et al. 2019). This was the first instance in which plant-associated bacteria in the environment were shown to use the T6SS to invade plant pests and replace intestinal endophytic bacteria. This observation was consistent with the previously reported phenomenon that both Salmonella typhimurium and Vibrio cholerae could use the T6SS to invade the host gut (Sana et al. 2016; Logan et al. 2018). The biological control effect of T6SS was also observed in Pseudomonas chlororaphis. Pseudomonas chlororaphis subsp. aureofaciens 30–84 is a plant growth-promoting bacterium that colonizes plant roots (Boak et al. 2022) and encodes two sets of T6SSs (T6SS-1 and T6SS-2). The existence of an arbitrary set of T6SSs in strain 30–84 can also affect the predation behavior of Tetrahymena thermophila, Dictyostelium discoideum, and Caenorhabditis elegans. The T6SS of strain 30–84 causes a stress response in predators, thereby protecting P. chlororaphis subsp. aureofaciens 30–84 from predation (Boak et al. 2022).
In conclusion, beneficial bacteria in plants can achieve biological control by employing the T6SS to inhibit plant pathogens and eukaryote bacterivores. These examples provide an important theoretical basis for plant-beneficial bacteria as biological control agents and suggest additional pathways for the development of new pathogenic bacteria and pest control strategies for crops, thus having significance in agricultural production.
Effects on the virulence of plant pathogens
The virulence of many plant-pathogenic bacteria is mediated by the type III secretion system (T3SS)(Meline et al. 2019; Yan et al. 2022). The T3SS can directly translocate toxic effector proteins to plant host cells (Jayaraman et al. 2020; Qiu et al. 2023). Although there are currently no reports on the direct injection of T6SS effector proteins into plant cells, many studies have shown that the loss of the T6SS reduces the virulence of plant pathogenic bacteria.
In Pseudomonas syringae pv. actinidiae M228, when the core genes tssM and tssJ of T6SS were deleted, the pathogenicity to kiwifruit branches decreased significantly, and phenotypes involving factors such as the bacterial competition ability, biofilm formation, hydrogen peroxide tolerance, and proteolytic activity were weakened (Wang et al. 2021b). This suggests that the T6SS plays an important role in pathogenic bacteria, probably via effects on bacterial competition, biofilm formation, and environmental adaptability (Wang et al. 2021b). The plant pathogen Burkholderia glumae BGR1, which causes bacterial panicle blight in rice, contains four T6SS gene clusters (T6SS group_1, T6SS group_2, T6SS group _ 4, and T6SS group_5). The virulence of tssD4 and tssD5 mutants to rice was decreased, indicating that T6SS group_4 and group_5, as eukaryotic targeting systems, are involved in the pathogenicity of bacteria to rice (Kim et al. 2020). Xanthomonas phaseoli pv. manihotis is the causal agent of cassava bacterial blight, an economically important disease in Africa and South America that causes losses that may reach 100% after three cassava production cycles. This local and systemic pathogen induces a wide combination of symptoms, including angular leaf spots, blight, wilting, dieback, gum exudation, and vascular necrosis (Montenegro Benavides et al. 2021). In one study, the T6SS core genes vgrG, clpV, tssM, and hcp in Xpm strain CIO151 were mutated, and the mutants were used to infect susceptible cassava plants. After 15 days of inoculation, the plant disease was alleviated compared with plants infected with wild-type bacteria, and the phenotype of the mutants was restored after genetic complementarity. The results suggest that these genes are necessary for the CIO151 infection of cassava plants, i.e., the T6SS mediates the pathogenicity of CIO151 to cassava plants (Montenegro Benavides et al. 2021).
The way in which the T6SS mediates bacterial pathogenic effects on plants is still unclear. It may be that bacteria use the T6SS to directly transfer toxic effector proteins into plant cells to produce symptoms. Alternately, the T6SS may indirectly affect virulence when the bacteria use the T6SS to gain a growth advantage in competition with other microbiota during plant colonization. A recent study performed transcriptome analysis of tssM mutants at the core of the type VI secretion system of Xanthomonas perforans and found that the T6SS affected the expression of global regulators such as csrA, rpoN, and pho, trigger signal cascade responses, and coordinate the expression of a series of virulence factors, stress response genes, and metabolic genes (Ramamoorthy et al. 2023). Therefore, the TssM of T6SS may be directly or indirectly involved in controlling the virulence of X. perforans. As such, the exact mechanism of the relationship between plant pathogenic bacteria T6SS and plant diseases remains to be clarified.
Regulation of the colonization of plant pathogens
Studies have shown that the T6SS plays an important role in the colonization of plant-associated bacteria (Lucero et al. 2022). Plant growth-promoting bacteria function through several mechanisms, including nitrogen fixation, the solubilization of insoluble soil phosphates, phytohormone production, and the prevention of diseases caused by pathogens (Ji et al. 2022). The T6SS of some bacteria can also inhibit the colonization of other bacteria on plants, thereby affecting plant growth.
A recent study (Lucero et al. 2022) identified strain J49 of Enterobacter sp. in peanuts as a strain that can colonize legumes and is an efficient phosphate solubilizer. The deletion mutation of the T6SS structural gene hcp in J49 significantly decreased the epiphytic and endophytic colonization rates of J49 in plant roots and aerial tissues, and the indexes related to plant colonization, including biofilm formation and polygalacturonase production, were lower than those of the wild-type strain (Lucero et al. 2022). The lack of T6SS function significantly weakened the colonization ability of J49 in legumes. In addition, a similar phenomenon was observed in Kosakonia sp. KO348 and P. fluorescens F113, in which mutation of the T6SS resulted in a phenotype with significantly decreased root colonization ability (Durán et al. 2021; Mosquito et al. 2022).
In contrast, the T6SS also appears to inhibit bacterial colonization in plants (Shidore et al. 2012). Shidore et al. found that in Azoarcus sp. BH72, when the same amount of wild-type strain BH72 and T6SS mutant BHazo3888 infected rice seedlings, the mutants showed stronger root colonization ability (Shidore et al. 2012). The same phenomenon was also observed in Acidovorax citrulli. Compared with the wild-type strain Aac5, the colony number of the strain with the mutant T6SS core gene hcp increased significantly after infecting watermelon cotyledons for 72 hours (Fei et al. 2022), indicating that T6SS was related to the colonization of A. citrulli (Kan et al. 2023).
In short, T6SS has two distinct effects on promoting and inhibiting the colonization of plant-associated bacteria in hosts. Therefore, we speculate that the inhibitory effect of T6SS on plant bacterial colonization may be related to the effector proteins secreted by the T6SS. Effector proteins secreted by the plant-associated bacterium T6SS trigger the local defense response of plants to kill wild-type strains. Since the T6SS mutant cannot secrete the effector protein, it will not trigger the local defense response of the plant, and thus the bacteria can effectively colonize the host plant (Shidore et al. 2012). However, this only explains how the T6SS inhibits bacterial colonization of plants, while the observation that the T6SS promotes bacterial colonization in plants may be explained by the inhibition of plant local defense responses by effector proteins. Therefore, if we can identify a T6SS effector protein that acts on plants and that has an effect on plant local defense systems, it will undoubtedly be a breakthrough in the field of T6SS research (Wang et al. 2023).
Influences on the symbiosis of Rhizobium and hosts
The T6SS exists widely in the genus Rhizobium and functions in both the promotion and inhibition of the symbiotic relationship between Rhizobium and legumes (Yang et al. 2022a).
Before the V. cholerae T6SS was identified, a gene encoding the T6SS had been identified in legume RBL5523, where the T6SS key gene impJ (tssK) mutant obtained via the insertion mutation of the Tn5 transposon caused pea plants to form more nodules and perform nitrogen fixation more effectively. This indicated that the T6SS could inhibit nodulation and nitrogen fixation in peas by inhibiting the symbiosis between Rhizobium and legumes (Bladergroen et al. 2003).
However, later studies have been more inclined to support the promoting effect of the T6SS on the symbiosis between Rhizobium and legumes. One study found that the plant dry weight and root nodule size produced by the mutation of the T6SS structural gene in Mim1 were lower than those of the wild-type strain. This was the first demonstration that the T6SS plays a promoting role in the symbiosis between Rhizobium and legumes (Salinero-Lanzarote et al. 2019). Similar phenomena have subsequently been reported in the rhizobia of many leguminous plants. For example, Paraburkholderia phymatum STM815 can nodulate a broad range of legumes, including the agriculturally important Phaseolus vulgaris (common bean) (Bellés-Sancho et al. 2023). P. phymatum harbors two T6SSs (T6SS-b and T6SS-3) in its genome. Differential gene expression analysis showed that T6SS-b could be activated in soil and the rhizosphere (Hug et al. 2021). Although no visible symbiotic phenotypes of the T6SS-b mutant were observed during the infection of germinated bean seeds, T6SS-b was expressed in the area of the root tip, suggesting that the expression of T6SS-b was caused by a component present on the tip of the root and indicating that T6SS was involved in early symbiosis with bean roots. R. leguminosarum LmicA16 is necessary for efficient nodulation in Lupinus micranthus and Lupinus angustifolius. After the mutation of the T6SS core structural gene vgrG in LmicA16, the mutant produced fewer nodules and smaller plants compared with the wild-type strain, and its competitiveness was weaker when co-inoculated with the wild-type strain. In addition, Tsb1 is a protein whose coding function is unknown but that contains a methyltransferase domain. The tsb1 mutant showed a phenotype between that of the wild type and the vgrG mutant. The effect of the tsb1 mutation suggests that the Tsb1 protein could be a possible T6SS effector that has a positive effect on the symbiosis between LmicA16 and Lupinus spp. (Tighilt et al. 2022).
Although no T6SS effector for the host cells of leguminous plants has been described, the beneficial effect of T6SS in symbiosis may be related to the attenuation of the defense response of the compatible host plant, as described by T3SS-dependent rhizobial effectors as effector-triggered susceptibility (Berrabah et al. 2019). This information may be significant for both ecological and agricultural research.
In conclusion, plant-associated bacteria improve their environmental adaptability and that of their plant hosts by employing the T6SS to compete with plant pathogens, inhibit plant pests, and influence colonization and symbiosis with their plant hosts. These examples provide an important theoretical basis concerning how some plant-associated bacteria resist biotic stresses in the environment, and thus provide further possibilities for developing new pathogen and pest control strategies to improve crop health.
T6SS confers resistance to abiotic stresses
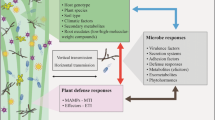
Plants have complex immune response strategies to defend against most microbes (Nishad et al. 2020). The immune defense response in plants includes pattern−triggered immunity (PTI) and effector-triggered immunity (ETI) (Spoel and Dong. 2012). The plant membrane pattern recognition receptors (PRRs) recognize microbe-associated molecular patterns (MAMPs) to trigger immune responses, thereby resulting in the accumulation of reactive oxygen species (ROS) (De Lorenzo et al. 2018). In addition, when infected by pathogens, plants will also produce organic acids or cause changes in the concentration of calcium and potassium ions inside and outside the cell, thus creating an acidic rhizosphere environment and a high osmotic gradient. Therefore, in addition to resistance to biotic stresses, bacteria are also resistant to various environmental stresses (Bhattacharyya et al. 2021) as well as some abiotic stresses released by the plant hosts, including oxidative stress, acid stress, and osmotic stress (Fig. 3) (Cui et al. 2015; Przepiora et al. 2019). There is some evidence that the T6SS plays a role in affecting the resistance of plant-associated bacteria to environmental stress and thus helps the cells to survive under adverse environmental conditions.
A model of the T6SS involved in the abiotic stress resistance of plant-associated bacteria. Under acid stress, acid-induced degradation of ExoRm may derepress ChvG to activate the T6SS of plant-associated bacteria by phosphorylation of the ChvI response regulator. Bacterial invasion of plants triggers plant immune defenses, leading to oxidative stress and osmotic stress. The expression of related genes in the T6SS gene cluster is altered and affects plant-associated bacterial resistance to stress
Acid stress
The intracellular pH plays an important role in regulating a variety of cellular processes, including cell growth, cell adhesion, signal transduction, and endocytosis (Wang et al. 2010). Bacteria infecting plant cells encounter a low pH in the apoplast (Jiang et al. 2016), and plants also release phenolic compounds as well as neutral and acidic sugars necessary to repair damaged tissue, thereby acidifying the rhizosphere to create a low pH environment that can limit the growth of pathogens (Winans. 1992; Subramoni et al. 2014). In response, pathogens have evolved multiple adaptation mechanisms, including using the T6SS in the stress response to low intracellular pH (Zhang et al. 2013; Qin et al. 2020; Yu et al. 2021).
Some plant-associated bacterial T6SSs can play a role in the intracellular low-pH stress response (Leonard et al. 2017). For example, when A. tumefaciens is near a suitable plant host in the rhizosphere, acidic conditions and plant-derived chemicals play important roles in activating its virulence (Winans. 1992; Yuan et al. 2008a). At the same time, after sensing the acidic features of the rhizosphere, A. tumefaciens exerts a conserved and signal-specific response to the plant host and adapts to the rhizosphere niche by regulating metabolism and cellular adaptation (Yuan et al. 2008a). A. tumefaciens can sense acidic pH through the ChvG/ChvI two-component system and activate the expression of several virulence factors, including the T6SS coding gene cluster (Yuan et al. 2008b; Subramoni et al. 2014). The periplasmic repressor ExoR negatively regulates the receptor kinase ChvG and reduces the expression and secretion activity of the T6SS. At acidic pH levels, the acid-induced degradation of periplasmic ExoR (ExoRm) may derepress ChvG to activate the T6SS through the phosphorylation of the ChvI response regulator (Wu et al. 2012). Due to the widespread existence of the ChvG/ChvI two-component system in α-Proteobacteria, the cascade regulation of the T6SS through ExoR-ChvG/ChvI may be a universal regulation mechanism in plant pathogens.
In addition to the ChvG/ChvI two-component system, there are other two-component systems capable of regulating the T6SS to help bacteria adapt to an acidic environment. OmpR is a well-characterized factor from the EnvZ/OmpR two-component system that regulates the expression of genes in response to changes in pH in the environment (Gerken et al. 2009). In Yersinia pseudotuberculosis, T6SS4 regulated by OmpR is essential for the bacteria to survive in acidic conditions, and OmpR can activate the T6SS to pump H+ out of cells to maintain the intracellular dynamic balance of pH. This may be a general strategy for pathogens to resist acidic conditions (Zhang et al. 2013). Alternatively, H3-T6SS, which is negatively regulated by OmpR in P. aeruginosa, and its secreted effector protein TepB, were able to aid in bacterial survival under acidic conditions (Yang et al. 2022b).
The regulatory mechanism of acid-induced T6SS is common in bacteria, and most feature two-component systems to respond to an external acidic environment. Whether acid-induced T6SS is a common regulatory mechanism in plant-associated bacteria in addition to the Exor-ChvG/ChvI cascade system is as yet unclear, as is the mechanism by which the T6SS participates in the response of plant bacteria against acid stress.
Oxidative stress
The production of ROS plays a key role in activating plant defense mechanisms (Torres. 2010; Khan et al. 2023). Through the activation of NADPH oxidases and peroxidases caused by the perception of pathogen-associated molecular patterns (PAMPs) by plants through PRRs, ROS are produced that in turn induce PTI-dependent basal defenses against invading pathogens. Following the perception of a PAMP, the produced ROS also act as toxins that can kill the pathogens (Park et al. 2013).
However, pathogens have evolved multiple mechanisms to evade host defenses by counteracting the deleterious effects of ROS (Wang et al. 2022). The T6SS is also involved in the evasion of host defenses. For example, Dickeya dadantii, a pathogenic bacterium that causes soft rot in many crops, employs the T6SS to combat oxidative stress and improve its environmental adaptability. In response to H2O2 treatment, D. dadantii expressed chemotactic genes to escape from the locally toxic environment and invade the plant intercellular space (Jiang et al. 2016). At the same time, the expression of several genes related to the type II, V, and VI secretion systems was downregulated (Jiang et al. 2016), indicating that the pathogen prioritized the allocation of its energy to survive rather than secrete late virulence factors. This phenomenon also shows that the T6SS of the plant-associated bacterium D. dadantii is involved in the process of resisting external oxidative stress through a special mechanism (Reverchon et al. 2016).
Unlike the T6SS, the use of the T3SS by plant pathogens to combat oxidative stress has been investigated, and studies have revealed antagonistic mechanisms. For example, the interaction between Ustilago maydis effector ROS burst interfering protein 1 (Rip1) and maize susceptibility factor lipoxygenase 3 (Zmlox3) results in the suppression of the ROS burst in several subcellular compartments of plant cells (Saado et al. 2022). Another way to resist oxidative stress is through the interaction between the effector protein and the target protein in the host. P. syringae effector HopAO1 targets lipooligosaccharide-specific reduced elicitation (LORE), a G-type lectin receptor-like kinase, to inhibit ROS bursts in Arabidopsis (Luo et al. 2020). Mechanistic studies of T3SS-secreted effector proteins against host oxidative stress also provide insights into the function of the T6SS. Although the mechanism of the T6SS in plant pathogens against oxidative stress in the environment is currently unclear, the T6SS in other bacteria is employed to resist oxidative stress by secreting effector proteins. For example, when Enterohemorrhagic E. coli (EHEC) colonizes the human intestine, macrophages will produce ROS to inhibit the bacteria. E. coli has a variety of different catalases to defend against oxidative stress (Poole. 2005; Imlay. 2008). A recent study showed that the T6SS of EHEC can secrete an effector protein KatN with catalase activity that can reduce the ROS level of host cells, thereby promoting the survival of EHEC in phagocytes (Wan et al. 2017). In addition, T6SS1 in Cupriavidus necator, which is regulated by the transcription factor Fur, secretes the lipopolysaccharide-binding effector TeoL to construct outer membrane vesicles in response to oxidative stress (Li et al. 2022). Therefore, in our future research, we will search for T6SS effector proteins examine their effects on host ROS levels, and further explore novel functions of T6SS in helping plant pathogens cope with oxidative stress.
Osmotic stress
The defense response of plants after infection promotes the influx of calcium ions to plant cells and the massive efflux of potassium ions in the apoplasts, resulting in osmotic pressure changes and cytoplasmic wall separation (Garcia-Brugger et al. 2006). Plant pathogens thus face osmotic stress when infecting plant cells. The tolerance to osmotic stress varies among bacteria. Differences in osmotic tolerance may be related to diverse adaptation strategies involving differences in osmotic adaptation mechanisms. These differences will affect the relative fitness of strains (Freeman et al. 2013). Studies have shown that the T6SS can improve bacterial osmotic adaptability (Gueguen et al. 2013; Guan et al. 2015), and this phenomenon also exists in plant bacteria.
Research has demonstrated that P. syringae B728a has strong osmotic tolerance. A whole-transcriptome analysis of B728a showed that 10 genes in the T6SS-Ι cluster, including clpV and tssM, are induced when the osmotic pressure increases (Freeman et al. 2013). According to the role of T6SS in antagonistic bacterial cell–cell interactions during protein transfer, the osmotic induction of the core T6SS machinery may be relevant to the interaction of B728a with other epiphytic organisms and may contribute to epiphytic fitness by antagonizing other microbes (Basler et al. 2013). In addition, the presence of tssM in X. perforans provides osmotic stress adaptation, facilitating the epiphytic colonization and growth of the pathogens (Liyanapathiranage et al. 2022). In the presence of a certain concentration of NaCl, compared with the tssM mutant, the wild-type strain X. perforans AL65 showed higher growth (Liyanapathiranage et al. 2022). This indicates that T6SS plays a role in adaptation to osmotic stress in the plant-associated bacterium X. perforans. In summary, T6SS is involved in the resistance of some plant-associated bacteria to osmotic stress. However, the ecological benefits of osmotic stress as a signal for T6SS induction are unclear.
The accumulation of compatible solutes is a strategy for bacteria to defend against high osmotic pressures in the environment (Kempf. 1998; Zeidler and Müller. 2019). The accumulation of specific compatible solutes in the bacterial cytoplasm also contributes to in-plant fitness in a strain-specific manner (Wang et al. 2021a). In addition, the responses of cells to osmotic pressure include potassium ion import. For example, bacteria use the two-component system KdpDE to regulate the transport of K+ by different proteins to maintain osmotic balance (Epstein. 2016; Schramke et al. 2016). Bacteria use the T6SS to transport metal ions (Lin et al. 2017; Si et al. 2017b; Han et al. 2019), and thus the T6SS in plant bacteria may also achieve osmotic adaptability by regulating the concentrations of intracellular and extracellular calcium and potassium ions. Future studies can take this as a starting point to investigate the mechanism by which the T6SS aids plant bacteria in resisting osmotic pressure.
In conclusion, the expression of the T6SS in plant-associated bacteria responds to external abiotic factors such as acid stress, oxidative stress, and osmotic stress. However, whether abiotic stress-induced T6SS is a common regulatory mechanism in plant-associated bacteria as well as the biological significance and application value of such regulation are important issues for future research.
Concluding remarks
The T6SS has been extensively studied as a versatile bacterial component. The T6SS was first described in V. cholerae, and subsequent research has largely focused on animal bacteria (Alteri and Mobley. 2016; Hernandez et al. 2020). However, the T6SS is also an extensive and very important system in plant-associated bacteria. This review summarizes the diverse functions of the T6SS in some common plant-associated bacteria, including the important role of the T6SS in resistance to biotic and abiotic stresses. In the resistance to biotic stress, the T6SS helps plant-associated bacteria improve their environmental adaptability and that of their plant hosts by competing with plant pathogens, inhibiting plant pests, and affecting colonization and symbiosis with plant hosts. However, in contrast to animal bacteria, T6SS effectors that can be directly secreted into plant cells have not yet been found, and the phenotypes of T6SSs that help plant-associated bacteria resist biological stresses have not been characterized. This is a research direction that requires further investigation in future studies.
There are few studies on plant-associated bacteria using the T6SS to resist abiotic stresses. The expression of T6SS-related genes has been shown to be altered under abiotic stress conditions. Although it has been speculated that this is due to the upregulation and downregulation of genes triggered by plant bacteria to resist environmental stress, the specific regulatory mechanisms remain unclear. The T6SS is regulated by multiple complex two-component systems in the cell, and some two-component systems can modulate the expression of the T6SS in response to abiotic stresses on plants, for example, the ExoR-ChvG/ChvI systems of A. tumefaciens that can activate the T6SS under acidic conditions. Further studies can target the regulation of the T6SS by abiotic stress-related intracellular components, explore the role of the T6SS in plant-associated bacterial resistance to abiotic stress, and expand the known functions of the T6SS in plant-associated bacterial stress resistance.
Plant-associated bacteria have been demonstrated to use the T6SS to compete with plant pathogens and adapt to abiotic stresses. This research provides a theoretical basis for improving the environmental adaptability of plant-associated bacteria and plant hosts. Therefore, it is crucial to explore the molecular mechanisms involving the T6SS in the interactions between plant-associated bacteria and their plant hosts. This may become an important direction for T6SS research, and it has potential application to the reduction of agricultural losses.
Availability of data and materials
Not applicable.
Abbreviations
- T6SS:
-
Type VI secretion system
- EI:
-
Effector protein-immune protein
- T3SS:
-
Type III secretion system
- ExoRm:
-
Periplasmic ExoR
- PTI:
-
Pattern−triggered immunity
- ETI:
-
Effector-triggered immunity
- PRRs:
-
Pattern recognition receptors
- MAMPs:
-
Microbe-associated molecular patterns
- ROS:
-
Reactive oxygen species
- PAMPs:
-
Pathogen-associated molecular patterns
- Rip1:
-
ROS burst interfering protein 1
- Zmlox3:
-
Maize susceptibility factor lipoxygenase 3
- LORE:
-
Lipooligosaccharide-specific reduced elicitation
References
Abo-Elyousr KAM, Abdel-Rahim IR, Almasoudi NM (2021) Native endophytic Pseudomonas putida as a biocontrol agent against common bean rust caused by Uromyces appendiculatus. J Fungi 7(9):745. https://doi.org/10.3390/jof7090745
Al-Karablieh N, Al-Shomali I, Al-Elaumi L, Hasan K (2022) Pseudomonas fluorescens NK4 siderophore promotes plant growth and biocontrol in cucumber. J Appl Microbiol 133(3):1414–1421. https://doi.org/10.1111/jam.15645
Alteri CJ, Mobley HLT (2016) The versatile type VI secretion system. Microbiol Spectr 4(2):337–356. https://doi.org/10.1128/microbiolspec.VMBF-0026-2015
Aschtgen MS, Bernard CS, De Bentzmann S, Lloubes R, Cascales E (2008) SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol 190(22):7523–7531. https://doi.org/10.1128/JB.00945-08
Basler M (2015) Type VI secretion system: secretion by a contractile nanomachine. Philos Trans R Soc Lond B Biol Sci 370(1679):20150021. https://doi.org/10.1098/rstb.2015.0021
Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ (2012) Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483(7388):182–186. https://doi.org/10.1038/nature10846
Basler M, Ho BT, Mekalanos JJ (2013) Tit-for-Tat type VI secretion counterattack during bacterial cell-cell interactions. Cell Microbiol 152(4):884–894. https://doi.org/10.1016/j.cell.2013.01.042
Bellés-Sancho P, Beukes C, James EK (2023) Nitrogen-Fixing Symbiotic Paraburkholderia Species Current Knowledge and Future Perspectives. Nitrogen 4(1):135–158. https://doi.org/10.3390/nitrogen4010010
Bendor L, Weyrich LS, Linz B, Rolin OY, Taylor DL, Goodfield LL, Smallridge WE, Kennett MJ, Harvill ET (2015) Type six secretion system of Bordetella bronchiseptica and adaptive immune. PloS One 10(10):e0140743. https://doi.org/10.1371/journal.pone.0140743
Bernal P, Allsopp LP, Filloux A, Llamas MA (2017) The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J 11(4):972–987. https://doi.org/10.1038/ismej.2016.169
Bernal P, Llamas MA, Filloux A (2018) Type VI secretion systems in plant-associated bacteria. Environ Microbiol 20(1):1–15. https://doi.org/10.1111/1462-2920.13956
Berrabah F, Ratet P, Gourion B (2019) Legume nodules massive infection in the absence of defense induction. Mol Plant Microbe Interact 32(1):35–44. https://doi.org/10.1094/MPMI-07-18-0205-FI
Bhattacharyya A, Pablo CHD, Mavrodi OV, Weller DM, Thomashow LS, Mavrodi DV (2021) Rhizosphere plant-microbe interactions under water stress. Adv Appl Microbiol 115:65–113. https://doi.org/10.1016/bs.aambs.2021.03.001
Bladergroen MR, Badelt K, Spaink HP (2003) Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol Plant Microbe Interact 16(1):53–64. https://doi.org/10.1094/MPMI.2003.16.1.53
Boak EN, Kirolos S, Pan H, Pierson LS 3rd, Pierson EA (2022) The type VI secretion systems in plant-beneficial bacteria modulate prokaryotic and eukaryotic interactions in the rhizosphere. Front Microbiol 13:843092. https://doi.org/10.3389/fmicb.2022.843092
Böck D, Medeiros JM, Tsao HF, Penz T, Weiss GL, Aistleitner K, Horn M, Pilhofer M (2017) In situ architecture, function, and evolution of a contractile injection system. Science 357(6352):713–717. https://doi.org/10.1126/science.aan7904
Bourigault Y, Dupont CA, Desjardins JB, Doan T, Bouteiller M, Le Guenno H, Chevalier S, Barbey C, Latour X, Cascales E, Merieau A (2023) Pseudomonas fluorescens MFE01 delivers a putative type VI secretion amidase that confers biocontrol against the soft-rot pathogen Pectobacterium atrosepticum. Environ Microbiol 25(11):2564–2579. https://doi.org/10.1111/1462-2920.16492
Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I (2009) Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. https://doi.org/10.1186/1471-2164-10-104
Cassan FD, Coniglio A, Amavizca E, Maroniche G, Cascales E, Bashan Y, de-Bashan LE (2021) The Azospirillum brasilense type VI secretion system promotes cell aggregation, biocontrol protection against phytopathogens and attachment to the microalgae Chlorella sorokiniana. Environmental Microbiol 23(10):6257–6274. https://doi.org/10.1111/1462-2920.15749
Chen L, Zou Y, Kronfl AA, Wu Y (2020) Type VI secretion system of Pseudomonas aeruginosa is associated with biofilm formation but not environmental adaptation. Microbiologyopen 9(3):e991. https://doi.org/10.1002/mbo3.991
Cherrak Y, Rapisarda C, Pellarin R, Bouvier G, Bardiaux B, Allain F, Malosse C, Rey M, Chamot-Rooke J, Cascales E, Fronzes R, Durand E (2018) Biogenesis and structure of a type VI secretion baseplate. Nat Microbiol 3(12):1404–1416. https://doi.org/10.1038/s41564-018-0260-1
Cherrak Y, Flaugnatti N, Durand E, Journet L, Cascales E (2019) Structure and activity of the type VI secretion system. Microbiol Spectr 7(4):PSIB-0031-2019. https://doi.org/10.1128/microbiolspec.PSIB-0031-2019
Coulthurst S (2019) The Type VI secretion system: a versatile bacterial weapon. Microbiol 165(5):503–515. https://doi.org/10.1099/mic.0.000789
Cui Z, Jin G, Li B, Kakar KU, Ojaghian MR, Wang Y, Xie G, Sun G (2015) Gene expression of type VI secretion system associated with environmental survival in acidovorax avenae subsp. avenae by principle component analysis. Int J Mol Sci 16(9):22008–22026. https://doi.org/10.3390/ijms160922008
De Lorenzo G, Ferrari S, Cervone F, Okun E (2018) Extracellular DAMPs in Plants and Mammals: Immunity Tissue Damage and Repair. Trends Immunol 39(11):937–950. https://doi.org/10.1016/j.it.2018.09.006
De Sousa BFS, Domingo-Serrano L, Salinero-Lanzarote A, Palacios JM, Rey L (2023) The T6SS-dependent effector Re78 of Rhizobium etli Mim1 benefits bacterial competition. Biology 12(5):678. https://doi.org/10.3390/biology12050678
Decoin V, Barbey C, Bergeau D, Latour X, Feuilloley MG, Orange N, Merieau A (2014) A type VI secretion system is involved in Pseudomonas fluorescens bacterial competition. PloS One 9(2):e89411. https://doi.org/10.1371/journal.pone.0089411
Durán D, Bernal P, Vazquez-Arias D (2021) Pseudomonas fluorescens F113 type VI secretion systems mediate bacterial killing and adaption to the rhizosphere microbiome. Sci Rep 11(1):5772. https://doi.org/10.1038/s41598-021-85218-1
El-Saadony MT, Saad AM, Soliman SM, Salem HM, Ahmed AI, Mahmood M, El-Tahan AM, Ebrahim AAM, Abd El-Mageed TA, Negm SH, Selim S, Babalghith AO, Elrys AS, El-Tarabily KA, AbuQamar SF (2022) Plant growth-promoting microorganisms as biocontrol agents of plant diseases: Mechanisms, challenges and future perspectives. Front Plant Sci 13:923880. https://doi.org/10.3389/fpls.2022.923880
Epstein W (2016) The KdpD Sensor Kinase of Escherichia coli Responds to Several Distinct Signals To Turn on Expression of the Kdp Transport System. J Bacteriol 198(2):212–220. https://doi.org/10.1128/JB.00602-15
Fei N, Ji W, Yang L, Yu C, Qiao P, Yan J, Guan W, Yang Y, Zhao T (2022) Hcp of the type VI secretion system (T6SS) in Acidovorax citrulli group II strain Aac5 has a dual role as a core structural protein and an effector protein in colonization, growth ability, competition, biofilm formation, and ferric Iron absorption. Int J Mol Sci 23(17):9632. https://doi.org/10.3390/ijms23179632
Felisberto-Rodrigues C, Durand E, Aschtgen MS, Blangy S, Ortiz-Lombardia M, Douzi B, Cambillau C, Cascales E (2011) Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog 7(11):e1002386. https://doi.org/10.1371/journal.ppat.1002386
Freeman BC, Chen C, Yu X (2013) Physiological and transcriptional responses to osmotic stress of two Pseudomonas syringae strains that differ in epiphytic fitness and osmotolerance. J Bacteriol 195(20):4742–4752. https://doi.org/10.1128/JB.00787-13
Gallegos-Monterrosa R, Coulthurst SJ (2021) The ecological impact of a bacterial weapon: microbial interactions and the Type VI secretion system. FEMS Microbiol Rev 45(6):fuab033. https://doi.org/10.1093/femsre/fuab033
Garcia-Brugger A, Lamotte O, Vandelle E, Bourque S, Lecourieux D, Poinssot B et al (2006) Early signaling events induced by elicitors of plant defenses. Mol Plant Microbe Interact 19(7):711–724. https://doi.org/10.1094/MPMI-19-0711
Gerken H, Charlson ES, Cicirelli EM, Kenney LJ, Misra R (2009) MzrA: a novel modulator of the EnvZ/OmpR two-component regulon. Mol Microbiol 72(6):1408–1422. https://doi.org/10.1111/j.1365-2958.2009.06728.x
Gu Q, Qiao J, Wang R, Lu J, Wang Z, Li P, Zhang L, Ali Q, Khan AR, Gao X, Wu H (2022) The role of pyoluteorin from Pseudomonas protegens Pf-5 in suppressing the growth and pathogenicity of Pantoea ananatis on maize. Int J Mol Sci 23(12):6431. https://doi.org/10.3390/ijms23126431
Guan J, Xiao X, Xu S, Gao F, Wang J, Wang T, Song Y, Pan J, Shen X, Wang Y (2015) Roles of RpoS in Yersinia pseudotuberculosis stress survival, motility, biofilm formation and type VI secretion system expression. J Microbiol 53(9):633–642. https://doi.org/10.1007/s12275-015-0099-6
Gueguen E, Durand E, Zhang XY, d’Amalric Q, Journet L, Cascales E (2013) Expression of aYersinia pseudotuberculosistype VI secretion system is responsive to envelope stresses through the OmpR transcriptional activator. PloS one 8(6):e66615. https://doi.org/10.1371/journal.pone.0066615
Han Y, Wang T, Chen G, Pu Q, Liu Q, Zhang Y, Xu L, Wu M, Liang H (2019) A Pseudomonas aeruginosa type VI secretion system regulated by CueR facilitates copper acquisition. PLoS Pathog 15(12):e1008198. https://doi.org/10.1371/journal.ppat.1008198
Hernandez RE, Gallegos-Monterrosa R, Coulthurst SJ (2020) Type VI secretion system effector proteins: Effective weapons for bacterial. Cell microbiol 22(9):e13241. https://doi.org/10.1111/cmi.13241
Ho BT, Dong TG, Mekalanos JJ (2014) A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15(1):9–21. https://doi.org/10.1016/j.chom.2013.11.008
Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7(1):25–37. https://doi.org/10.1016/j.chom.2009.12.007
Huang FC, Chi SF, Chien PR, Liu YT, Chang HN, Lin CS, Hwang HH (2021) Arabidopsis RAB8A, RAB8B and RAB8D proteins interact with several RTNLB proteins and are involved in the Agrobacterium tumefaciens infection process. Plant Cell Physiol 62(10):1572–1588. https://doi.org/10.1093/pcp/pcab112
Hug S, Liu Y, Heiniger B, Bailly A, Ahrens CH, Eberl L, Pessi G (2021) Differential expression of Paraburkholderia phymatum type VI secretion systems (T6SS) suggests a role of T6SS-b in early symbiotic interaction. Front Plant Sci 12:699590. https://doi.org/10.3389/fpls.2021.699590
Hug S, Heiniger B, Bolli K, Paszti S, Eberl L, Ahrens CH, Pessi G (2023) Paraburkholderia sabiae uses one type VI secretion system (T6SS-1) as a powerful weapon against notorious plant pathogens. Microbiology Spectr 11(4):e0162223. https://doi.org/10.1099/mic.0.2007/006585-0
Imlay JA (2008) Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem 77:755–776. https://doi.org/10.1146/annurev.biochem.77.061606.161055
Jayaraman J, Yoon M, Applegate ER, Stroud EA, Templeton MD (2020) AvrE1 and HopR1 from Pseudomonas syringae pv. actinidiae are additively required for full virulence on kiwifruit. Mol Plant Pathol 21(11):1467–1480. https://doi.org/10.1111/mpp.12989
Ji C, Tian H, Wang X, Song X, Ju R, Li H, Gao Q, Li C, Zhang P, Li J, Hao L, Wang C, Zhou Y, Xu R, Liu Y, Du J, Liu X (2022) Bacillus subtilis HG-15, a halotolerant rhizoplane bacterium, promotes growth and salinity tolerance in wheat (Triticum aestivum). Biomed Res Int:9506227. https://doi.org/10.1155/2022/9506227
Jiang X, Zghidi-Abouzid O, Oger-Desfeux C, Hommais F, Greliche N, Muskhelishvili G, Nasser W, Reverchon S (2016) Global transcriptional response of Dickeya dadantii to environmental stimuli relevant to the plant infection. Environ Microbiol 18(11):3651–3672. https://doi.org/10.1111/1462-2920.13267
Kan Y, Zhang Y, Lin W, Dong T (2023) Differential plant cell responses to Acidovorax citrulli T3SS and T6SS reveal an effective strategy for controlling plant-associated pathogens. mBio Online ahead of print:e0045923. https://doi.org/10.1128/mbio.00459-23
Kapitein N, Bonemann G, Pietrosiuk A, Seyffer F, Hausser I, Locker JK, Mogk A (2013) ClpV recycles VipA/VipB tubules and prevents non-productive tubule formation to ensure efficient type VI protein secretion. Mol Microbiol 87(5):1013–1028. https://doi.org/10.1111/mmi.12147
Kempf B, Bremer E (1998) Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol 170(5):319–330. https://doi.org/10.1007/s002030050649
Kempnich MW, Sison-Mangus MP (2020) Presence and abundance of bacteria with the Type VI secretion system in a coastal environment and in the global oceans. PloS One 15(12):e0244217. https://doi.org/10.1371/journal.pone.0244217
Khan M, Ali S, Al Azzawi TNI, Saqib S, Ullah F, Ayaz A, Zaman W (2023) The Key Roles of ROS and RNS as a Signaling Molecule in Plant-Microbe Interactions. Antioxidants (Basel) 12(2). https://doi.org/10.3390/antiox12020268
Kim N, Kim JJ, Kim I, Mannaa M, Park J, Kim J, Lee HH, Lee SB, Park DS, Sul WJ, Seo YS (2020) Type VI secretion systems of plant-pathogenic Burkholderia glumae BGR1 play a functionally distinct role in interspecies interactions and virulence. Mol Plant Pathol 21(8):1055–1069. https://doi.org/10.1111/mpp.12966
Le Goff M, Vastel M, Lebrun R, Mansuelle P, Diarra A, Grandjean T, Triponney P, Imbert G, Gosset P, Dessein R, Garnier F, Durand E (2022) Characterization of the Achromobacter xylosoxidans type VI secretion system and its implication in cystic fibrosis. Front Cell Infect Microbiol 12:859181. https://doi.org/10.3389/fcimb.2022.859181
Leonard S, Hommais F, Nasser W, Reverchon S (2017) Plant-phytopathogen interactions: bacterial responses to environmental and plant stimuli. Environ Microbiol 19(5):1689–1716. https://doi.org/10.1111/1462-2920.13611
Li C, Zhu L, Wang D, Wei Z, Hao X, Wang Z, Li T, Zhang L, Lu Z, Long M, Wang Y, Wei G, Shen X (2022) T6SS secretes an LPS-binding effector to recruit OMVs for exploitative competition and horizontal gene transfer. ISME J 16(2):500–510. https://doi.org/10.1038/s41396-021-01093-8
Lin J, Cheng J, Chen K, Guo C, Zhang W, Yang X, Ding W, Ma L, Wang Y, Shen X (2015) The icmF3 locus is involved in multiple adaptation- and virulence-related characteristics in Pseudomonas aeruginosa PAO1. Front Cell Infect Microbiol 5:70. https://doi.org/10.3389/fcimb.2015.00070
Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, Wei G, Qian PY, Luo ZQ, Shen X (2017) A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun 8:14888. https://doi.org/10.1038/ncomms14888
Lin J, Xu L, Yang J, Wang Z, Shen X (2021) Beyond dueling: roles of the type VI secretion system in microbiome modulation, pathogenesis and stress resistance. Stress Biology 1:1–12. https://doi.org/10.1007/s44154-021-00008-z
Liu M, Wang H, Liu Y, Tian M, Wang Z, Shu RD, Zhao MY, Chen WD, Wang H, Wang H, Fu Y (2023) The phospholipase effector Tle1Vc promotes Vibrio cholerae virulence by killing competitors and impacting gene expression. Gut microbes 15(1):2241204. https://doi.org/10.1080/19490976.2023.2241204
Liyanapathiranage P, Jones JB, Potnis N (2022) Mutation of a single core gene, tssM, of type VI secretion system of Xanthomonas perforans influences virulence, epiphytic survival, and transmission during pathogenesis on tomato. Phytopathology 112(4):752–764. https://doi.org/10.1094/PHYTO-02-21-0069-R
Logan SL, Thomas J, Yan J, Baker RP, Shields DS, Xavier JB, Hammer BK, Parthasarathy R (2018) The Vibrio cholerae type VI secretion system can modulate host intestinal mechanics to displace gut bacterial symbionts. Proc Natl Acad Sci U S A 115:E3779–E3787. https://doi.org/10.1073/pnas.1720133115
Lucero CT, Lorda GS, Ludueña LM, Nievas F, Bogino P, Angelini JG, Ambrosino ML, Taurian T (2022) Participation of type VI secretion system in plant colonization of phosphate solubilizing bacteria. Rhizosphere 24:100582. https://doi.org/10.1016/j.rhisph.2022.100582
Luo X, Wu W, Liang Y, Xu N, Wang Z, Zou H, Liu J (2020) Tyrosine phosphorylation of the lectin receptor-like kinase LORE regulates plant immunity. EMBO J 39(4):e102856. https://doi.org/10.15252/embj.2019102856
Ma LS, Hachani A, Lin JS, Filloux A, Lai EM (2014) Agrobacterium tumefaciens deploys a superfamily of type VI secretion DNase effectors as weapons for interbacterial competition in planta. Cell Host Microbe 16(1):94–104. https://doi.org/10.1016/j.chom.2014.06.002
Marchi M, Boutin M, Gazengel K, Rispe C, Gauthier JP, Guillerm-Erckelboudt AY, Lebreton L, Barret M, Daval S, Sarniguet A (2013) Genomic analysis of the biocontrol strain Pseudomonas fluorescens Pf29Arp with evidence of T3SS and T6SS gene expression on plant roots. Environ Microbiol Rep 5(3):393–403. https://doi.org/10.1111/1758-2229.12048
Meline V, Delage W, Brin C, Li-Marchetti C, Sochard D, Arlat M, Rousseau C, Darrasse A, Briand M, Lebreton G, Portier P, Fischer-Le Saux M, Durand K, Jacques MA, Belin E, Boureau T (2019) Role of the acquisition of a type 3 secretion system in the emergence of novel pathogenic strains of Xanthomonas. Molecular Plant Pathology 20(1):33–50. https://doi.org/10.1111/mpp.12737
Montenegro Benavides NA, Alvarez B A, Arrieta-Ortiz ML, Rodriguez-R LM, Botero D, Tabima JF, Castiblanco L, Trujillo C, Restrepo S, Bernal A (2021) The type VI secretion system of Xanthomonas phaseoli pv. manihotis is involved in virulence and in vitro motility. BMC Microbiol 21(1):14. https://doi.org/10.1186/s12866-020-02066-1
Mosquito S, Bertani I, Licastro D, Compant S, Myers MP, Hinarejos E, Levy A, Venturi V (2022) In planta colonization and role of T6SS in two rice Kosakonia endophytes. Mol Plant Microbe Interact 33(2):349–363. https://doi.org/10.1094/MPMI-09-19-0256-R
Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312(5779):1526–1530. https://doi.org/10.1126/science.1128393
Nanjani S, Soni R, Paul D, Keharia H (2022) Genome analysis uncovers the prolific antagonistic and plant growth-promoting potential of endophyte Bacillus velezensis K1. Gene 836:146671. https://doi.org/10.1016/j.gene.2022.146671
Nguyen VS, Logger L, Spinelli S, Legrand P, Huyen Pham TT, Nhung Trinh TT, Cherrak Y, Zoued A, Desmyter A, Durand E, Roussel A, Kellenberger C, Cascales E, Cambillau C (2017) Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex. Nat Microbiol 2:17103. https://doi.org/10.1038/nmicrobiol.2017.103
Nishad R, Ahmed T, Rahman VJ, Kareem A (2020) Modulation of Plant Defense System in Response to Microbial Interactions. Front Microbiol 11:1298. https://doi.org/10.3389/fmicb.2020.01298
Park SY, Choi J, Lim SE, Lee GW, Park J, Kim Y, Kong S, Kim SR, Rho HS, Jeon J, Chi MH, Kim S, Khang CH, Kang S, Lee YH (2013) Global expression profiling of transcription factor genes provides new insights into pathogenicity and stress responses in the rice blast fungus. PLoS Pathog 9(6):e1003350. https://doi.org/10.1371/journal.ppat.1003350
Pietrosiuk A, Lenherr ED, Falk S, Bönemann G, Kopp J, Zentgraf H, Sinning I, Mogk A (2011) Molecular basis for the unique role of the AAA+ chaperone ClpV in type VI protein secretion. J Biol Chem 286(34):30010–30021. https://doi.org/10.1074/jbc.M111.253377
Poole LB (2005) Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys 433(1):240–254. https://doi.org/10.1016/j.abb.2004.09.006
Phttps://doi.org/10.1007/s10658-019-01902-z (2019) Effects of stressful physico-chemical factors on the fitness of the plant pathogenic bacterium Dickeya solani. Eur J Plant Pathol 156(2):519–535. https://doi.org/10.1007/s10658-019-01902-z
Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103(5):1528–1533. https://doi.org/10.1073/pnas.0510322103
Qin L, Wang X, Gao Y, Bi K, Wang W (2020) Roles of EvpP in Edwardsiella piscicida-Macrophage Interactions. Front Cell Infect Microbiol 10:53. https://doi.org/10.3389/fcimb.2020.00053
Qiu H, Wang B, Huang M, Sun X, Yu L, Cheng D, He W, Zhou D, Wu X, Song B, Tang N, Chen H (2023) A novel effector RipBT contributes to Ralstonia solanacearum virulence on potato. Mol Plant Pathol 24(8):947–960. https://doi.org/10.1111/mpp.13342
Ramamoorthy S, Pena M, Ghosh P, Liao YY, Paret ML, Jones JB, Potnis N (2023) Transcriptome profiling of type VI secretion system core gene tssM mutant of Xanthomonas perforans highlights regulators controlling diverse functions ranging from virulence to metabolism. bioRxiv 13:548846. https://doi.org/10.1101/2023.07.13.548846
Reverchon S, Muskhelisvili G, Nasser W (2016) Virulence program of a bacterial plant pathogen the Dickeya model. Prog Mol Biol Transl Sci 142:51–92. https://doi.org/10.1016/bs.pmbts.2016.05.005
Ripcke K, Romero Picazo D, Vichare S, Unterweger D, Dagan T, Huelter NF (2023) T6SS-mediated competitive exclusion among Pantoea agglomerans associated with plants. bioRxiv 17:533219. https://doi.org/10.1101/2023.03.17.533219
Ruhe ZC, Low DA, Hayes CS (2020) Polymorphic toxins and their immunity proteins: diversity, evolution, and mechanisms of delivery. Annu Rev Microbiol 74:497–520. https://doi.org/10.1146/annurev-micro-020518-115638
Saado I, Chia KS, Betz R, Alcântara A, Pettkó-Szandtner A, Navarrete F, D'Auria JC, Kolomiets MV, Melzer M, Feussner I, Djamei A (2022) Effector-mediated relocalization of a maize lipoxygenase protein triggers susceptibility to Ustilago maydis. Plant Cell 34(7):2785–2805. https://doi.org/10.1093/plcell/koac105
Salinero-Lanzarote A, Pacheco-Moreno A, Domingo-Serrano L, Durán D, Ormeño-Orrillo E, Martínez-Romero E, Albareda M, Palacios JM, Rey L (2019) The type VI secretion system of Rhizobium etli Mim1 has a positive effect in symbiosis. FEMS Microbiol Ecol 95(5):054. https://doi.org/10.1093/femsec/fiz054
Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut (2016) Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci U S A 113(34):E5044–5051. https://doi.org/10.1073/pnas.1608858113
Santoriello F, Pukatzki S (2023) Type VI secretion systems: Environmental and Intra-host competition of Vibrio cholerae. Adv Exp Med Biol 1404:41–63. https://doi.org/10.1007/978-3-031-22997-8_3
Savary S, Willocquet L (2020) Modeling the impact of crop diseases on global food security. Annu Rev Phytopathol 58:313–341. https://doi.org/10.1146/annurev-phyto-010820-012856
Schramke H, Tostevin F, Heermann R, Gerland U, Jung K (2016) A Dual-Sensing Receptor Confers Robust Cellular Homeostasis. Cell Rep 16(1):213–221. https://doi.org/10.1016/j.celrep.2016.05.081
Shidore T, Dinse T, Öhrlein J, Becker A, Reinhold-Hurek B (2012) Transcriptomic analysis of responses to exudates reveal genes required for rhizosphere competence of the endophyte Azoarcus sp. strain BH72. Environmental Microbiol 14(10):2775–2787. https://doi.org/10.1111/j.1462-2920.2012.02777.x
Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG (2013) PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500(7462):350–353. https://doi.org/10.1038/nature12453
Si M, Zhao C, Burkinshaw B, Zhang B, Wei D, Wang Y, Dong TG, Shen X (2017) Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc Natl Acad Sci U S A 114(11):E2233–E2242. https://doi.org/10.1073/pnas.1614902114
Si M, Wang Y, Zhang B, Zhao C, Kang Y, Bai H, Wei D, Zhu L, Zhang L, Dong TG, Shen X (2017) The type VI secretion system engages a redox-regulated dual-functional heme transporter for zinc acquisition. Cell Rep 20(4):949–959. https://doi.org/10.1016/j.celrep.2017.06.081
Spoel SH, Dong X (2012) How do plants achieve immunity? Defence without specialized immune cells. Nat Rev Immunol 12(2):89–100. https://doi.org/10.1038/nri3141
Stubbendieck RM, Vargas-Bautista C, Straight PD (2016) Bacterial communities: interactions to scale. Front Microbiol 7:1234. https://doi.org/10.3389/fmicb.2016.01234
Subramoni S, Nathoo N, Klimov E, Yuan ZC (2014) Agrobacterium tumefaciens responses to plant-derived signaling molecules. Front Plant Sci 5:322. https://doi.org/10.3389/fpls.2014.00322
Tang JY, Bullen NP, Ahmad S (2018) Diverse NADase effector families mediate interbacterial antagonism via the type VI secretion system. J Biol Chem 293(5):1504–1514. https://doi.org/10.1074/jbc.RA117.000178
Tang MX, Pei TT, Xiang Q, Wang ZH, Luo H, Wang XY, Fu Y, Dong T (2022) Abiotic factors modulate interspecies competition mediated by the type VI secretion system effectors in Vibrio cholerae. ISME J 16(7):1765–1775. https://doi.org/10.1038/s41396-022-01228-5
Taylor NMI, van Raaij MJ, Leiman PG (2018) Contractile injection systems of bacteriophages and related systems. Mol Microbiol 108(1):6–15. https://doi.org/10.1111/mmi.13921
Tighilt L, Boulila F, De Sousa BFS, Giraud E, Ruiz-Argüeso T, Palacios JM, Imperial J, Rey L (2022) The Bradyrhizobium Sp. LmicA16 type VI secretion system is required for efficient nodulation of Lupinus Spp. Microb Ecol 84(3):844–855. https://doi.org/10.1007/s00248-021-01892-8
Torres MA (2010) ROS in biotic interactions. Physiol Plant 138(4):414–429. https://doi.org/10.1111/j.1399-3054.2009.01326.x
Trunk K, Peltier J, Liu YC, Dill BD, Walker L, Gow NAR, Stark MJR, Quinn J, Strahl H, Trost M, Coulthurst SJ (2018) The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol 3(8):920–931. https://doi.org/10.1038/s41564-018-0191-x
Turner TR, James EK, Poole PS (2013) The plant microbiome. Genome Biol 14(6):209. https://doi.org/10.1186/gb-2013-14-6-209
Vacheron J, Péchy-Tarr M, Brochet S, Heiman CM, Stojiljkovic M, Maurhofer M, Keel C (2019) T6SS contributes to gut microbiome invasion and killing of an herbivorous pest insect by plant-beneficial Pseudomonas protegens. ISME J 13(5):1318–1329. https://doi.org/10.1038/s41396-019-0353-8
Wan B, Zhang Q, Ni J, Li S, Wen D, Li J, Xiao H, He P, Ou HY, Tao J, Teng Q, Lu J, Wu W, Yao YF (2017) Type VI secretion system contributes to Enterohemorrhagic Escherichia coli virulence by secreting catalase against host reactive oxygen species (ROS). PLoS Pathog 13(3):e1006246. https://doi.org/10.1371/journal.ppat.1006246
Wang R, Yu C, Yu F (2010) Molecular fluorescent probes for monitoring pH changes in living cells. TrAC Trends in Anal Chem 29(9):1004–1013. https://doi.org/10.1016/j.trac.2010.05.005
Wang J, Brodmann M, Basler M (2019) Assembly and subcellular localization of bacterial type VI secretion systems. Annu Rev Microbiol 73:621–638. https://doi.org/10.1146/annurev-micro-020518-115420
Wang H, Yang Z, Swingle B, Kvitko BH (2021) AlgU, a conserved sigma factor regulating abiotic stress tolerance and promoting virulence in Pseudomonas syringae. Mol Plant Microbe Interact 34(4):326–336. https://doi.org/10.1094/MPMI-09-20-0254-CR
Wang N, Han N, Tian R, Chen J, Gao X, Wu Z, Liu Y, Huang L (2021) Role of the type VI secretion system in the pathogenicity of Pseudomonas syringae pv. actinidiae, the causative agent of kiwifruit bacterial canker. Front Microbiol 12:627785. https://doi.org/10.3389/fmicb.2021.627785
Wang Y, Pruitt RN, Nurnberger T, Wang Y (2022) Evasion of plant immunity by microbial pathogens. Nat Rev Microbiol 20(8):449–464. https://doi.org/10.1038/s41579-022-00710-3
Wang J, Yin R, Yang J, Cheng J, Lin J (2023) Research progress in the function of type VI secretion system in plant-associated bacteria. Acta Microbiologica Sinica 63(07):2573–2594. https://doi.org/10.13343/j.cnki.wsxb.20220817
Whitney JC, Chou S, Russell AB, Biboy J, Gardiner TE, Ferrin MA, Brittnacher M, Vollmer W, Mougous JD (2013) Identification, structure, and function of a novel type VI secretion peptidoglycan glycoside hydrolase effector-immunity pair. J Biol Chem 288(37):26616–26624. https://doi.org/10.1074/jbc.M113.488320
Winans SC (1992) Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol Rev 56:12–31. https://doi.org/10.1128/mr.56.1.12-31.1992
Wu CF, Lin JS, Shaw GC, Lai EM (2012) Acid-induced type VI secretion system is regulated by ExoR-ChvG/ChvI signaling cascade in Agrobacterium tumefaciens. PLoS Pathog 8(9):e1002938. https://doi.org/10.1371/journal.ppat.1002938
Wu CF, Santos MNM, Cho ST, Chang HH, Tsai YM, Smith DA, Kuo CH, Chang JH, Lai EM (2019) Plant-pathogenic Agrobacterium tumefaciens strains have diverse type VI effector-immunity pairs and vary in in-planta competitiveness. Mol Plant Microbe Interact 32(8):961–971. https://doi.org/10.1094/MPMI-01-19-0021-R
Yan Y, Wang Y, Yang X, Fang Y, Cheng G, Zou L, Chen G (2022) The MinCDE cell Division system participates in the regulation of type III secretion system (T3SS) genes, bacterial virulence, and motility in Xanthomonas oryzae pv. oryzae. Microorganisms 10(8):1549. https://doi.org/10.3390/microorganisms10081549
Yang X, Long M, Shen X (2018) Effector-Immunity pairs provide the T6SS nanomachine its offensive and defensive capabilities. Molecules 23(5):1009. https://doi.org/10.3390/molecules23051009
Yang J, Lan L, Jin Y, Yu N, Wang D, Wang E (2022) Mechanisms underlying legume–rhizobium symbioses. J Integr Plant Biol 64(2):244–267. https://doi.org/10.1111/jipb.13207
Yang Y, Pan D, Tang Y, Li J, Zhu K, Yu Z, Zhu L, Wang Y, Chen P, Li C (2022) H3–T6SS of Pseudomonas aeruginosa PA14 contributes to environmental adaptation via secretion of a biofilm-promoting effector. Stress Biology 2(1):55. https://doi.org/10.1007/s44154-022-00078-7
Yu KW, Xue P, Fu Y, Yang L (2021) T6SS mediated stress responses for bacterial environmental survival and host adaptation. Int J Mol Sci 22(2):478. https://doi.org/10.3390/ijms22020478
Yuan ZC, Liu P, Saenkham P, Kerr K, Nester EW (2008) Transcriptome profiling and functional analysis of Agrobacterium tumefaciens reveals a general conserved response to acidic conditions (pH 5.5) and a complex acid-mediated signaling involved in Agrobacterium-plant interactions. J Bacteriol 190(2):494–507. https://doi.org/10.1128/JB.01387-07
Yuan ZC, Haudecoeur E, Faure D, Kerr KF, Nester EW (2008) Comparative transcriptome analysis of Agrobacterium tumefaciens in response to plant signal salicylic acid, indole-3-acetic acid and gamma-amino butyric acid reveals signalling cross-talk and Agrobacterium–plant co-evolution. Cell Microbiol 10(11):2339–2354. https://doi.org/10.1111/j.1462-5822.2008.01215.x
Zeidler S, Müller V (2019) Coping with low water activities and osmotic stress in Acinetobacter baumannii significance, current status and perspectives. Environ Microbiol 21(7):2212–2230. https://doi.org/10.1111/1462-2920.14565
Zhang W, Wang Y, Song Y, Wang T, Xu S, Peng Z, Lin X, Zhang L, Shen X (2013) A type VI secretion system regulated by OmpR in Yersinia pseudotuberculosis functions to maintain intracellular pH homeostasis. Environ Microbiol 15(2):557–569. https://doi.org/10.1111/1462-2920.12005
Zhang D, Yan M, Niu Y, Liu X, van Zwieten L, Chen D, Pan G (2016) Is current biochar research addressing global soil constraints for sustainable agriculture? Agricult Ecosyst Environ 226:25–32. https://doi.org/10.1016/j.agee.2016.04.010
Zhang J, Li W, Huang X, Li D, Liu X, Jiang X, Li X (2023) Genomic and transcriptomic analysis revealed new insights into the influence of key T6SS genes hcp and vgrG on drug resistance and interbacterial competition in Klebsiella pneumoniae. bioRxiv 16:540999. https://doi.org/10.1101/2023.05.16.540999
Zoued A, Durand E, Brunet YR, Spinelli S, Douzi B, Guzzo M, Flaugnatti N, Legrand P, Journet L, Fronzes R, Mignot T, Cambillau C, Cascales E (2016) Priming and polymerization of a bacterial contractile tail structure. Nature 531(7592):59–63. https://doi.org/10.1038/nature17182
Acknowledgments
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (32070103), the Qinchuang Yuan “Scientist + Engineer” Team Construction Project of Shaanxi Province (2023KXJ-019), the Regional Development Talent Project of the “Special Support Plan” of Shaanxi Province 2020-44, a grant from the Outstanding Young Talent Support Plan of the Higher Education Institutions of Shaanxi Province 2018-111, and the Youth Innovation Team of Shaanxi Universities 2022-943.
Author information
Authors and Affiliations
Contributions
J.L. conceptualized the article; R.Y. performed the literature search and wrote the manuscript, and R.Y. and J.C. prepared the figures and tables. All authors contributed to the article and have approved the submitted version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors agree to publish.
Competing interests
The authors declare no competing financial interests.
Additional information
Handling editor: Lei Xu.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yin, R., Cheng, J. & Lin, J. The role of the type VI secretion system in the stress resistance of plant-associated bacteria. Stress Biology 4, 16 (2024). https://doi.org/10.1007/s44154-024-00151-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s44154-024-00151-3