Abstract
The bacterium Rhodococcus erythropolis MI2 uses 4,4´-dithiodibutyric acid (DTDB) as carbon source to synthesize polythioesters (PTE). The first step for the production of PTE using DTDB is catalyzed by an NADH:flavin oxidoreductase (nox) as it was previously shown in our laboratory, and the second step is catabolized by a putative luciferase-like monooxygenase (Llm). In the current study, experiments were carried out to identify the function of Llm. Hence, the llm gene, which encodes for the Llm protein, was amplified from the genomic DNA of MI2 using polymerase chain reaction and expressed in Escherichia coli BL21 cells. Protein purification was done using His Spin Trap affinity columns. Enzyme assay was carried out using the purified protein and p-coumaric acid as substrate giving a specific activity of 1.6 U/mg protein for the purified Llm. The responsible gene (llm) was deleted in the genome of MI2, and a single deletion mutant was subsequently generated. Finally, growth of the wild-type strain (MI2) and the mutant strain (MI2Δllm) were compared using DTDB or succinate as carbon sources. Whereas the wild type was successfully grown with DTDB or succinate, the llm-negative mutant exhibited low grow with DTDB although it grows very well with succinate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rhodococcus are aerobic, Gram-positive bacteria with high G + C contents, which play an important role in the degradation of a wide range of xenobiotic substances [1, 2]. Often, they harbor large linear plasmids in addition to their large chromosomes which can be involved in the transformation of various organic compounds [3, 4]. They are present in soils polluted with crude oil or xenobiotic compounds, and they are frequently used in bioremediation processes [5]. They play a major role in the desulfurization of fossil fuels [6], and in the commercial production of acrylamide [7]. R. erythropolis ATCC 4277 and R. erythropolis CCM2595 are involved in the desulfurization of dibenzothiophene and in the bioremediation of aromatic compounds, respectively [8, 9]. Polythioesters (PTE) are sulfur-containing nonbiodegradable polymers, accumulated in the cytoplasm of few bacteria under special conditions [10]. These persistent polymers have very interesting properties and could be used in industrial applications [11,12,13]. PTE were produced in Ralstonia eutropha, recombinant Escherichia coli and Advenella mimigardefordensis [11, 14,15,16,17].
4,4´-Dithiodibutyric acid (DTDB), is a white, solid organic disulfide used as an alternative monolayer for the manufacture of protein chips, which are applied in various sugars estimation by Raman spectroscopy and cyclic voltammetry [18, 19]. Although DTDB is a structural analogue of homocysteine, its biodegradation is observed only in few bacteria [20]. Cultivation of bacteria with DTDB is easy, because it is non-toxic substrate and available in the market. Khairy et al. [1] reported on the DTDB catabolism in Rhodococcus spp and identified the genes. The first step in DTDB catabolism is cleavage of DTDB into two molecules of 4-mercaptobutyric acid (4 MB) by an NADH:flavin oxidoreductase (nox) [1, 21, 22]. 4 MB is a toxic compound because of the availability of free sulfhydryl group which inhibits bacterial growth, hence no reports were available about production of PTE using 4 MB as carbon source [23]. Khairy et al. [1, 21, 22] proposed that the second step in DTDB catabolism is the oxidation of 4 MB into 4-oxo-4-sulfanylbutyric acid by a putative luciferase-like monooxygenase (Llm); however, this has not been proved, yet. Hence, the objective of the present study is to evaluate the function of Llm in the catabolism of DTDB, more specifically to find out the oxidation capability of the substrate, p-coumaric acid. To test this, the responsible gene (llm) was amplified from genomic DNA of MI2, and heterologously expressed in E. coli BL21 cells using both pET19b and pET23a expression vectors. After protein purification, the enzyme activity was monitored in a spectrometric assay. Moreover, the llm gene was deleted in the genome of MI2, and growth of the wild-type strain and the mutant strain (MI2Δllm) were monitored using DTDB or succinate.
Materials and methods
Bacteria
The bacteria and plasmids used are listed in Table 1. Strain MI2 and deletion mutant of MI2 were grown at 30 °C in LB [24] or in mineral salts medium (MSM) [25] under aerobic condition. E. coli strains were grown in LB medium at 37 °C under aerobic condition. DTDB was used as carbon source at a concentration of 30 mM in MSM with the medium pH adjusted to 7.0.
Protein expression
DNA isolation and PCR amplification
To clone and express the llm gene in E. coli BL21 cells, and to generate the deletion mutant with lacking llm gene, total DNA (gDNA) was isolated from cells of strain M12. Approximately 20 ml of LB medium in a 100 ml of Erlenmeyer flask was inoculated with a single colony of MI2 and incubated overnight at 30 °C in a shaking incubator. The NucleoSpin® Tissue Kit (Macherey–Nagel GmbH and Co. KG) was used to purify the gDNA. MI2 is a Gram-positive bacterium and difficult to lyse; hence, pre-lysis was done with the bacterial pellet obtained after centrifugation by incubating at 37 °C for 2 h with 20 mg of lysozyme and 450 μl of Tris buffer (NaCl—0.44 g; EDTA—0.93 g; Tris HCl—0.24 g; water—100 ml). Proteinase K (25 μl) was added to the mixture to stop DNA degradation by DNAses and then incubated at 56 °C for 2 h. DNA was isolated according to the manufacturer instructions using this mixture.
The llm gene was amplified from the total genomic DNA by polymerase chain reaction (PCR). The primers were designed with the SeqBuilder (DNASTAR) and synthesized by MWG-Biotech AG (Germany). Only one forward primer, but two different reverse primers (one for pET19b vector and one for pET23a vector) were used for gene amplification (Table 2). The primers were designed in a way that the obtained fragments can later be cut by restriction enzymes and ligated into the multiple cloning site (MCS) of pET19b and pET23a vectors. PCR done with Phusion high-fidelity DNA polymerase or with Biomix (Bioline, UK) using the thermal cycler peqSTAR 2X/Gradient from PEQLAB Biotechnologie GmbH. The genome of Rhodococcus is GC-rich; therefore, the GC buffer was used. The dimethyl sulfoxide (DMSO) binds to the DNA on the cytosine overhangs and changes its conformation, making the DNA more susceptible to heat denaturation. This simplifies the annealing of the primers to the DNA. An agarose gel electrophoresis was carried out to check the size of the amplified genes. If the fragments were correct, a Gel Extraction Kit (PEQLAB Biotechnologie GmbH, Germany) was used to purify the PCR products from agarose gel. After the purification, the concentrations were measured with the NanoPhotometer® N60. Purified products were used for cloning in various vectors.
Restriction digestion and ligation
Restriction digestion was done according to Khairy et al. [1] to generate the llm gene fragments which can be ligated into suitable vectors. Genes and plasmids to be ligated were digested with the same restriction enzymes. The reaction mixture contains buffer, vector with gene, restriction enzyme and water were incubated at 37 °C for 1–2 h and then purified using the Gel Extraction Kit to remove the restriction enzymes, ligase and buffers or the enzymes were inactivated by a heat shock treatment (65 °C, 5 min). Purified DNA fragments were ligated, the ligation mixture contains the buffer, T4 DNA ligase, vector and gene. Ligation mixtures were incubated for 30 min at room temperature, and then ligase was deactivated by a heat shock at 65 °C for 5 min. All the chemicals, buffers and enzymes used for ligation were purchased from Thermo Fisher Scientific.
Cloning
To check whether Llm is actually involved in the DTDB catabolism or not, we need to perform the enzyme assay. Cloning of the llm gene was done as mentioned below. First, llm was amplified from the genomic DNA of MI2 strain using PCR, intermediate cloning was done using the PCR product and the vector pJET1.2/blunt (CloneJET Kit, Thermo Scientific) based on the supplier’s instructions. The blunt end PCR products were ligated directly into the vector which is an easy and reliable method because the amplified genes safely ligate and replicate in the plasmid. T4 ligase ligates the fragments by making a new phosphodiester linkage between the respective ends. Expression vectors pET19b and pET23a were used for the T7 promoter based heterologous expression. The llm gene was cut out from the hybrid plasmid pJET1.2::llm with appropriate restriction endonucleases and purified with the peqGOLD Gel Extraction kit and then cloned into both, the pET19b and the pET23a vectors. Both vectors were used, because pET19b contains the N-terminal His-tag, and pET23a contains the C-terminal His-tag. For the pET23a, llm gene was digested with HindIII and NdeI, for the pET19b llm gene was digested with BamHI and NdeI. Restriction digested products were checked on agarose gel, bands were observed at expected size, i.e., llm gene (930 bp), pET23a vector (3600 bp), and the pET19b vector (5700 bp) indicated that the restriction digestion worked properly. Ligation into the pET vectors occurred due to complementary single stranded sticky ends between vector and fragments. The ligation products pET19b::llm or pET23a::llm were then transformed into E. coli Top10 competent cells. Successful transformants were selected based on LB ampicillin, the hybrid plasmids were isolated, analyzed by sequencing, and confirmed that no point mutations were observed. For sequencing, samples were sent to Eurofins Genomics in a total volume of 17 μl. Obtained sequence results were monitored by Seqlab software (Göttingen, Germany) to check the gene sequence. The verified clones were transformed into BL21 competent cells for the expression of protein.
Purification of the protein
Heterologous expression of llm gene in E. coli BL21 cells was accomplished by cultivation in an auto-inductive (ZYP-5052) medium or in LB medium. IPTG (0.4 mM) was added to the LB medium to induce the cells. ZYP medium (100 ml) supplemented with ampicillin was inoculated with 1% of preculture. Initially cells were cultivated at 37 °C for 4 h and then at 20 °C. The cultures were grown on a rotary shaker at 130 rpm for about 18 h. Pellet was collected by centrifugation, washed twice with sterile saline, and resuspended in 50 mM Tris–HCl buffer (pH 7.4), containing 20 mM imidazole and subsequently interrupted by 4 times passage through a French press (100 × 106 Pa). Centrifugation (9000×g, 4 °C, 60 min) was done to obtain the supernatants containing soluble protein fractions from crude extracts. The supernatant was used for Llm protein purification. All buffers were used as suggested by the His Spin Trap affinity columns (GE Healthcare, United Kingdom) providers for the purification of histidine-tagged fusion proteins. Tris–HCl buffer with 20 mM imidazole was used for column equilibration. Buffers with 50 mM Tris–HCl and various imidazole concentrations (40 and 100 mM) were used for the first and second washing steps to achieve the better purity. Finally, elution was carried out using an elution buffer which contains 500 mM imidazole. At the end, protein was transferred to sodium phosphate buffer (pH 7.4) after removing imidazole in Vivaspin 500 columns (Sartorius AG, Germany). In most cases, bacteria showed higher protein expression with ZYP medium than by inducing with IPTG. Protein concentrations were measured using a spectral photometer (Thermo Spectronic, USA) by measuring the OD at 595 nm as mentioned [10].
SDS-PAGE
Protein samples and SDS loading buffer were mixed together, boiled and used for SDS-PAGE analysis. SDS in combination with a reducing agent (DTT or β-mercaptoethanol) will denatures the proteins. Gels were casted with 11.5% separating gel and 4% collecting gel. From each fraction, samples containing 40 µg protein (crude extract, lysate, flow through, washing fractions) were loaded onto the gel. For elution fractions, 5 μg of protein were loaded on the gel. A color prestained protein standard (10 μl) with broad range from 11 to 245 kDa from NEB was also loaded. The gel was first run at 40 mA until the samples from the collecting gel were moved to the separating gel and then allowed to run at 80–100 mA. The gels were incubated for 5–7 min in SDS-PAGE staining solution, de-stained overnight with 10% acetic acid, and scanned with the Epson scanner and the images were captured.
Enzyme test
Enzyme assay was done according to the protocol of Haghbeena and Tan [26]. 1 ml of reaction mixture contains 50 mmol/l potassium phosphate buffer (pH 7. 4), 0.1 mmol/l NADPH, 0.3 µmol/l FMN, 5 mmol/l p-coumaric acid (substrate) and 10 µg Llm (enzyme) were taken in a polystyrene cuvette and the enzyme activity was measured at 30 °C by monitoring the decrease in the absorbance at 288 nm due to the oxidation of p-coumaric acid to caffeic acid. Activities are shown in units (U), where 1 U is the activity of the enzyme that converts 1 µmol of substrate per minute. Controls, without substrate, without enzyme, and without cofactor were also performed. The effect of pH on Llm activity was studied at 30 °C in a pH range of 5–9. Sodium acetate (pH 4.0–5.0), MES (pH 6.0), potassium phosphate (pH 7.4), Tris/Cl (pH 8.0), and MOPS (pH 9.0) buffers at 50 mmol/l concentrations were used. The influence of temperature on the reaction rate was analyzed in the range of 20–80 °C using phosphate buffer. The specific enzyme activity was also calculated. Originally, the oxidation of 4 MB to 4-oxo-4-sulfanylbutyric acid could be evaluated. But, 4 MB is a highly toxic due to the sulfhydryl group and will prevent the growth of bacteria [23, 27]; hence, its oxidation is a crucial step for the growth of bacteria. The main function of Llm is oxidation of the substrates; hence, the oxidation of p-coumaric acid to caffeic acid was studied. Unlike 4 MB, the substrate p-coumaric acid is non-toxic compound and promotes bacterial growth.
Deletion mutants
The deletion mutant was prepared by deleting the gene llm in MI2 for the better understanding of DTDB metabolic pathway. The suicide plasmid technique was used to obtain defined deletion mutants [28]. Deletion of llm was accomplished by cloning the upstream and downstream flanking regions of the gene into the XbaI restriction site of the suicide plasmid pJQ200mp18Tc. Upstream (484 bp), downstream (950 bp) fragments of llm, and pJQ vector were digested with the same restriction enzyme (XbaI), and ligated to produce a 1434-bp fragment. The resulting gene replacement plasmid (pJQ200mp18Tc::Δllm) was multiplied in E. coli TOP10 cells. To verify that the flank was inserted into the vector, a colony PCR was done. PCR products were loaded on agarose gel and the correct transformants were identified. Plasmid purification, restriction digestion was done for the selected two colonies and sent for sequencing. MI2 competent cells were electroporated with 50 μg of plasmid DNA and streaked on LB agar tetracycline plates. The plates were incubated for several days at 30 °C and the formation of clones was observed.
Preparation of competent cells
Competent cells of MI2 were prepared as mentioned in the protocol [29, 30]. Precultures were prepared by adding a loopful of MI2 cells in 10 ml of LB medium and incubated overnight at 30 °C under shaking conditions. The main culture was prepared by adding 5% preculture in 50 ml of LB medium in the 300-ml Erlenmeyer flask and incubated overnight under shaking at 30 °C. When the OD600 reached between 0.9 and 1.2, the cells were centrifuged, and the resultant pellet was resuspended in 20 ml of ice-cold sterile Milli Q water. Centrifugation was done again, the supernatant was discarded and the resultant pellet washed twice with 10 ml of cold Milli Q sterile water containing 10% glycerol. After final centrifugation, the pellet was resuspended in 1.25 ml of milli Q water with 10% glycerol, aliquoted (100 μl) in Eppendorf tubes and stored at − 70 °C until further use.
Electroporation conditions
The suicide vector ligated to us/ds flanks of llm was transformed into MI2 cells through electroporation. 20 μl of plasmid DNA (0.5–1 μg) together with 100 μl of the competent cells was incubated on ice for 30 min. Subsequently, the reaction mixture was pipetted into an ice-cold 2 mm electroporation cuvette (Bio-Budget Technologies GmbH) and electroporated at 2.5 kV, 400 Ω and 25 μF using the Gene Pulser Xcell™ electroporation system (Bio-Rad Laboratories GmbH). For each time, two pulses lasting between 4 and 10 ms were performed. The cells were then added to 900 μl SOC medium and taken in 15-ml falcon tubes and incubated under shaking at 30 °C for 4 h, and then centrifuged for two min at 9000 rpm. The obtained pellet was resuspended in 200 μl of SOC medium, spreading was done with 100 μl of the cell suspension on two LB agar tetracycline plates and were incubated at 30 °C for several days.
Growth experiments
Strain MI2 was able to use succinate or DTDB as carbon and energy source on solid plates. Therefore, it was cultivated for 48 h in liquid cultures at 30 °C in MSM. MSM was supplemented with 30 mM DTDB or 50 mM sodium succinate as carbon source. Erlenmeyer flasks with baffles were used for growth of liquid cultures to guarantee an optimal oxygen supply, and the flasks were incubated on a rotary shaker at 120 rpm. Both, the wild-type and the mutant strains, were investigated in growth experiments. Controls without bacterial cells were also used. Optical density was measured at various time periods using a Klett Summerson photometer to evaluate the growth pattern. Growth experiments were performed in triplicates, the results were presented as average and standard deviation. The experimental steps conducted in this study are summarized in Fig. 1.
Results and discussion
Heterologous expression and purification of Llm
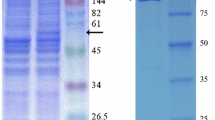
After the isolation of genomic DNA from the wild-type strain MI2, llm was amplified through PCR. llm has the length of 930 bp, the size of llm was confirmed by agarose gel electrophoresis. Before heterologous expression in E. coli BL21 cells, gene sequences were verified using Seqlab software, and it was confirmed that no mutations had occurred in the gene sequence. Higher protein concentration was obtained with pET23a hybrid vector than pET19b hybrid vector. E. coli BL21 cells, which harbor pET23a::llm showed higher protein concentrations in ZYP medium (crude extract: 1.99 mg/ml, lysate: 2.05 mg/ml, flow through: 1.79 mg/ml, wash1: 1.25 mg/ml, wash2: 0.79 mg/ml, and elution: 0.82 mg/ml). Similarly, cells gown in LB medium also showed significant protein concentration (crude extract: 1.76 mg/ml, lysate: 1.78 mg/ml, flow through: 1.42 mg/ml, wash1: 0.98 mg/ml, wash2: 0.61 mg/ml, and elution: 0.78 mg/ml). Similar amounts of protein concentrations were observed in the crude extract and lysate. Relatively large amounts of protein in the flow through were observed, as the unbound protein is washed directly from the column. The amount of protein in the washing steps decreased, because the unbound or weakly bound protein will be washed out and only pure protein will remain. Different fractions obtained during the protein purification were loaded on the SDS-PAGE gel to verify the purity and the molecular weight. The target protein (Llm) had the molecular weight of 34 kDa through the His-tag with linker (about 1 kD) the expected size is about 35 kDa. SDS gel image of various fractions are shown in Fig. 2. Initially, many bands were observed in elution buffer, but later washing conditions were optimized to get rid of non-specific bands.
SDS-PAGE gel image of the purified Llm protein. The protein was heterologously expressed in E. coli BL21 cells harboring the llm gene. Cells were grown in auto-induction medium, and protein purification was done through His Spin Trap affinity columns. Except elution, amounts of 40 μg protein were applied onto the gel from each sample. Elution fractions comprising 5 μg protein were applied onto the gel. M marker, CE crude extract, Lys lysate, FT flow through, W1 wash1, W2 wash2, E elution
Initially, double digestions were done to cut llm from the vector, but it was not successful. Hence, further attempts were made with sequential digestion until successful results were obtained. During protein purification, low amounts of soluble proteins were produced when the cultures were incubated at 30 °C, it might be due to the formation of inclusion bodies which usually consist of incompletely folded insoluble and inactive proteins. This is a common phenomenon during protein overexpression. Hence, cells were first incubated at 37 °C until the OD600 reached values between 0.4 and 0.6, then induced with IPTG and further incubated at 20 °C. The low temperature leads to slow growth of cells and subsequently slow down the synthesis of proteins. By this method formation of inclusion bodies can be avoided; hence, we grew the cultures at 20 °C in this study and obtained significantly better results. According to literature reports, protein solubility depends on protein folding, and protein folding may be depending on C- or N-terminal His-tag. Length of C- or N-terminal tags shows influence on protein folding and solubility. Small tags with up to 12 amino acids (His-tag) and large with more than 60 amino acids can be available [31, 32]. In most cases, small tags do not cause conformational changes, but it is possible because not only tag size but also charge and hydrophobicity will exert an influence on protein folding. The His-tag is charged and small, sometimes causes poor solubility of the protein, hence some researchers use larger hydrophilic tag [32]. For example, the glutathione-S-transferase (GST) tag is slightly larger but known to increase the solubility and stability of the fusion proteins [33]. The advantages of using His tags are: (i) elution is simple, (ii) many purification protocols are reported, (iii) tags are small and need not to be removed enzymatically. A disadvantage is the high imidazole concentrations to be used for elution Furthermore, elution may cause precipitation, so the imidazole must be eliminated through dialysis [34].
Enzyme analysis
Enzyme activity was measured by monitoring the decrease in the absorbance at 288 nm due to the oxidation of p-coumaric acid to caffeic acid (Fig. 3). Originally, the oxidation of 4 MB to 4-oxo-4-sulfanylbutyric acid could be evaluated, but 4 MB is a highly toxic and expensive. The main function of Llm is oxidation of the substrates, hence the oxidation of p-coumaric acid to caffeic acid was studied. Unlike 4 MB, the substrate p-coumaric acid is not toxic and inexpensive. Decrement in the absorption of substrate at 288 nm was observed, highest decrement in the absorption was observed when co-factors were used along with enzyme and substrate. The effect of pH on Llm activity was studied at 30 °C in a pH range of 4.0–9.0. Higher enzyme activity was achieved at pH 7.5 using potassium phosphate buffer. Phosphate buffer is more heat-stable and is suitable for measuring the enzyme activity at higher temperatures. The optimum specific activity was obtained at 40 °C, conspicuous activities were recorded at 55–75 °C. Llm was specific for NADPH and FMN, higher activity was recorded after adding both co-factors to the reaction mixture. The specific enzyme activity of purified Llm was 1.6 U/mg protein. Llm enzyme activity measured in this study was in accordance with the proteomics studies previously conducted by Khairy et al. [22]. They reported that Llm, which is encoded by one of the 126 monooxygenase encoding genes in MI2, exhibited a three times enhanced expression level with DTDB than succinate in the proteome analysis.
Measurement of Llm enzyme activity at 30 °C by monitoring the decrease in the absorbance at 288 nm due to the oxidation of p-coumaric acid to caffeic acid. 1 ml of reaction mixture contains 50 mmol/l potassium phosphate buffer (pH 7. 4), 0.1 mmol/l NADPH, 0.3 µmol/l FMN, 5 mmol/l p-coumaric acid (substrate) and 10 µg Llm (enzyme). E Enzyme, S Substrate, CoF Cofactor
Flavoprotein monooxygenases are involved in a variety of biological processes ranging from lignin degradation to the biosynthesis of natural products and detoxification of toxic compounds [35]. They use FMN or FAD as cofactor to activate dioxygen and they catalyze the incorporation of one atom of O2 into a substrate and the reduction of the other oxygen atom to water. Literature reports denoted the functions and properties of monooxygenases. Zhou et al. [36] found that a flavin-dependent monooxygenase converted the naphthoquinone chromophore of rifamycin S into benzo-γ-pyrone and also linearized rifamycin SV through phenolic hydroxylation. Pal and Sengupta [37] denoted that Rhodococcus alkane-1-monooxygenase showed maximum activity at alkaline pH. Zhu et al. [38] shown that an FAD-dependent monooxygenase catalyzes oxygen insertion into an amide bond to form the key 2H-tetrahydro-4,6-dioxo-1,2-oxazine ring in alchivemycin, which is a highly oxidized polycyclic compound with potent antimicrobial activity.
Luciferases constitute a well-studied group of flavoproteins, observed in bioluminescence organisms such as fungi, glowworms, fish, and bacteria. Bacterial luciferases utilize O2 and FMNH2 to oxidize long chain aliphatic aldehydes into a fatty acid and emit light. LLMs conduct various oxidations, such as Baeyer–Villiger (BV) oxidation, epoxidation, sulfoxidation, and alkane hydroxylation [36]. The bacterial luciferase from Vibrio harveyi is a flavin-containing monooxygenase that oxidizes aldehyde to a carboxylic acid while emitting visible light [39]. Alali et al. [40] reported that FMN-dependent LLM involved in the production of rishirilides which are a group of PKS II secondary metabolites produced by Streptomyces bottropensis. Li et al. [41] found that four LLM class flavin-dependent oxidoreductase coding genes. Mascotti et al. [42] found the well-known bacterial FMN-dependent luciferases such as LuxB and PDB: 1luc.
Deletion mutants and growth experiments
The electroporation of the suicide vector harboring llm into MI2 strain did not yield clones at the beginning, which might be either due to cells were not competent enough for DNA uptake or the electroporation process did not work properly. Optimization of electroporation conditions, plasmid, and gene concentrations were not successful. Stecker [43] reported that transformation efficiency is low in R. erythropolis, and sometimes it is difficult requiring several attempts to get the clones on plates. Various attempts were made to introduce the DNA into the cells via electroporation rather than conjugation, or transduction, or protoplast transformation [44, 45]. We assume that the Llm enzyme will be involved in the oxidation of 4 MB to 4-oxo-4-sulfanylbutyric acid in DTDB catabolism, and it was proven, because the purified Llm protein oxidized the substrate coumaric acid. Therefore, the responsible gene (llm) was deleted in the genome of MI2 and single deletion mutant was subsequently generated. The reason to generate mutant was to find out whether the protein/enzyme encoded by this llm gene is really involved in the DTDB catabolism or not by comparing the growth behavior of this mutant with the wild type using DTDB as carbon source.
Growth experiments were conducted after having the llm gene mutant (MI2∆llm) available. The wild-type strain MI2 was able to use succinate or DTDB as sole carbon sources, but the deletion mutant showed less growth on solid agar plates with DTDB, than with succinate as sole carbon sources. Thereafter, the wild-type and mutant strains were separately cultivated for 48 h in liquid cultures at 30 °C in MSM containing 50 mM succinate or 30 mM DTDB as carbon sources. Erlenmeyer flasks with baffles were used in liquid cultures, and the flasks were incubated on a rotary shaker at 120 rpm. In liquid growth experiments, deletion mutant MI2∆llm showed lower growth with DTDB than with succinate (Fig. 4). When the cells were grown with DTDB, the wild type entered the exponential phase after 18 h cultivation. Clear difference in the growth of the mutant and the wild-type strains were noticed from this point. After 18 h growth, the stationary phase starts with wild type at 30 h with OD of 489 KU, whereas the mutant showed OD of 324 KU at 30 h. At 48 h, cells of the mutant with OD of 278 KU, were unable to reach the optical density of the wild type (OD of 428 KU). Contrary to this, the wild type and mutant showed almost similar growth pattern, and no significant difference in growth was observed when they were cultivated with succinate. Wild type and mutant entered the exponential phase after 12 h with an OD value of 195 KU. After that both strains showed rapid growth, and the OD value reached to 570 KU at 48 h cultivation time and then both strains entered the decline phase. In the growth experiments, the wild type was successfully grown with DTDB or succinate, but the mutant showed lower growth with DTDB thus indicating that Llm encoded by llm might be involved in the catabolism of DTDB.
Growth curves of the wild type (R. erythropolis MI2) and the mutant (R. erythropolis MI2Δllm) using DTDB or succinate as sole carbon source. Both carbon sources were separately added in liquid MSM to a concentration of 30 mM or 50 mM, respectively. Triplicate experiments were done, and error bars are shown. KU indicates Klett Units
DTDB degradation and metabolic pathway
DTDB is an important substrate for the synthesis of poly(4 MB); hence, the catabolism of DTDB in MI2 must be unraveled (Fig. 5). Studies related with DTDB degradation and metabolites identification were not performed in the present study, because it was already proved several times in our laboratory using GC–MS analysis [1, 20,21,22]. The initial step in DTDB degradation is the reduction of the disulfide bond, which results in the formation of two molecules of 4 MB [20]. This reaction is catalyzed by Nox as it was confirmed by the enzymatic in vitro analysis [21]. 4 MB is a highly toxic mercaptoalkanoic acid and seems to be more toxic than other mercaptoalkanoates [23, 27] (Fig. 5). The oxidation of 4 MB to 4-oxo-4-sulfanylbutyric acid is the second step in DTDB catabolism. The high toxicity of 4 MB is caused by the sulfhydryl group and inhibits bacterial growth [23]; hence, its oxidation is crucial for the bacterial growth. 4-Oxo-4-sulfanylbutyric acid was detected in the wild-type grown cultures, last step in DTDB catabolism is the transformation of 4-oxo-4-sulfanylbutyric acid into succinic acid [20]. MI2 strains were grown with MSM containing 0.5% sodium gluconate as carbon source during the studies conducted by Khairy et al. [1] in our laboratory. The cells were harvested and transferred to MSM containing sodium gluconate plus 30 mM DTDB. After 72 h, MI2 utilized all the DTDB. 4 MB, 4-oxo-4-sulfanylbutyric acid, succinic acid, and volatile hydrogen sulfide were identified which supported the results described by Wübbeler et al. [20]. Later, Khairy et al. [22] also reported about the utilization of DTDB using the wild-type MI2. They used 30 mM DTDB as initial carbon source in MSM, and the availability of remaining DTDB was calculated at different time intervals. They reported that strain MI2 was unable to utilize the DTDB until 12 h, after that utilization of DTDB started and almost 50% of DTDB was utilized within 36 h.
A schematic representation of the catabolism of DTDB in R. erythropolis MI2. Dotted line arrows show the expected pathway for Poly(4-mercaptobutyrate) production using DTDB as carbon source. 1: NADH-flavin oxidoreductase (Nox); 2: Luciferase-like monooxygenase (Llm); 3: Sulfide quinone oxidoreductase (Sqr)
Conclusions
In continuation of the work carried out by Khairy et al. [21], this study focused on further investigations of the postulated metabolic pathway of DTDB in R. erythropolis MI2. To achieve this, llm was successfully amplified from genomic DNA of MI2, heterologously expressed in E. coli BL21 cells using pET19b or pET23a vectors, and the protein was purified. Purity and molecular weight of the protein were confirmed by SDS-PAGE analysis. It was shown that Llm oxidized the substrate, p-coumaric acid. In growth experiments, the cells of the wild type were grown very well with DTDB or succinate whereas the mutant strain, MI2∆llm showed only very little grow with DTDB even though it grew very well with succinate indicating that the llm gene is involved in the DTDB catabolism. The results obtained from these studies proved that Llm is involved in the oxidation of substrates and more specifically involved in the catabolism of DTDB and further production of polythioesters.
Availability of data and material
The authors declare that data supporting the findings of this study are available within the article and its supplementary information files.
Code availability
Not applicable.
References
Khairy H, Wübbeler JH, Steinbüchel A. Biodegradation of the organic disulfide 4,4´-dithiodibutyric acid by Rhodococcus spp. Appl Env Microbiol. 2015;81:8294–306.
Larkin MJ, Kulakov LA, Allen CCR. Biodegradation by members of the genus Rhodococcus: biochemistry, physiology and genetic adaptation. Adv Appl Microbiol. 2006;59:1–29.
Alvarez HM, editor. Microbiology monographs, vol 16. Biology of Rhodococcus. Berlin: Springer; 2010.
van der Geize R, Dijkhuizen L. Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr Opin Microbiol. 2004;7:255–61.
Bell KS, Philp JC, Aw DWJ, Christofi N. The genus Rhodococcus. J Appl Microbiol. 1998;85:195–210.
Matsui T, Noda K, Tanaka Y, Maruhashi K, Kurane R. Recombinant Rhodococcus sp. strain T09 can desulfurize DBT in the presence of inorganic sulfate. Curr Microbiol. 2002;45:240–4.
Komeda H, Hori Y, Kobayashi M, Shimizu S. Transcriptional regulation of the Rhodococcus rhodochrous J1 nitA gene encoding a nitrilase. Proc Nat Acad Sci USA. 1996;93:10572–7.
Maass D, de Oliveira D, de Souza AAU, Smagu S. Biodesulfurization of a system containing synthetic fuel using Rhodococcus erythropolis ATCC 4277. Appl Biochem Biotechnol. 2014;174:2079–85.
Szoköl J, Rucká L, Šimcíková M, Halada P, Nešvera J, Pátek M. Induction and carbon catabolite repression of phenol degradation genes in Rhodococcus erythropolis and Rhodococcus jostii. Appl Microbiol Biotechnol. 2014;98:8267–79.
Reddy MV, Steinbüchel A. 3,3-Thiodipropionic acid (TDP), a possible precursor for the synthesis of polythioesters: identification of TDP transport proteins in Variovorax paradoxus TBEA6. Appl Microbiol Biotechnol. 2021;105:3733–43.
Lütke-Eversloh T, Kawada J, Marchessault RH, Steinbüchel A. Characterization of biological polythioesters: physical properties of novel copolymers synthesized by Ralstonia eutropha. Biomacromol. 2002;3:159–66.
Lütke-Eversloh T, Steinbüchel A. Microbial polythioesters. Macromol Biosci. 2004;4:165–74.
Wübbeler JH, Steinbüchel A. New pathways for bacterial polythioesters. Curr Opi Biotechnol. 2014;29:85–92.
Lütke-Eversloh T, Bergander K, Luftmann H, Steinbüchel A. Identification of a new class of biopolymer: bacterial synthesis of a sulfur containing polymer with thioester linkages. Microbiology. 2001;147:11–9.
Lütke-Eversloh T, Bergander K, Luftmann H, Steinbüchel A. Biosynthesis of poly(3-hydroxybutyrate-co-3-mercaptobutyrate) as a sulfur analogue to poly(3-hydroxybutyrate) (PHB). Biomacromol. 2001;2:1061–5.
Thakor N, Lütke-Eversloh T, Steinbüchel A. Application of the BPEC pathway for large scale biotechnological production of poly(3-mercaptopropionate) by recombinant Escherichia coli, including a novel in situ isolation method. Appl Env Microbiol. 2005;71:835–41.
Xia Y, Wübbeler JH, Qi Q, Steinbüchel A. Employing a recombinant strain of Advenella mimigardefordensis for biotechnical production of homopolythioesters from 3,3´-dithiodipropionic acid. Appl Env Microbiol. 2012;78:3286–97.
Jang LS, Keng HK. Development and characterization of 4,4´-dithiodibutyric acid as a monolayer for protein chips. Sensors Mat. 2006;18:367–80.
Kanayama N, Kitano H. Interfacial recognition of sugars by boronic acid-carrying self-assembled monolayers. Langmuir. 2000;16:577–83.
Wübbeler JH, Bruland N, Wozniczka M, Steinbüchel A. Biodegradation of the xenobiotic organic disulphide 4,4´-dithiodibutyric acid by Rhodococcus erythropolis strain MI2 and comparison to the microbial utilization of 3,3´-dithiodipropionic acid and 3,3´-thiodipropionic acid. Microbiology. 2010;156:1221–33.
Khairy H, Wübbeler JH, Steinbüchel A. The NADH:flavin oxidoreductase Nox from Rhodococcus erythropolis MI2 is the key enzyme of 4,4´-dithiodibutyric acid degradation. Lett Appl Microbiol. 2016;63:434–41.
Khairy H, Meinert C, Wübbeler JH, Poehlein A, Daniel R, Voigt B, Riedel K, Steinbüchel A. Genome and proteome analysis of Rhodococcus erythropolis MI2: elucidation of the 4,4´-dithiodibutyric acid catabolism. PLoS One. 2016;11:1–22.
Lütke-Eversloh T, Steinbüchel A. Novel precursor substrates for polythioesters (PTE) and limits of PTE biosynthesis in Ralstonia eutropha. FEMS Microbiol Lett. 2003;221:191–6.
Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1989.
Schlegel HG, Gottschalk G, Bartha RV. Formation and utilization of poly-β-hydroxybutyric acid by Knall gas bacteria (Hydrogenomonas). Nature. 1961;191:463–5.
Haghbeena K, Tan EW. Dirreddect spectrophotometric assay of monooxygenase and oxidase activities of mushroom tyrosinase in the presence of synthetic and natural substrates. Anal Biochem. 2003;3:23–32.
Lütke-Eversloh T, Steinbüchel A. Polythioesters. In: Matsumura S, Steinbüchel A, editors. Biopolymers, vol. 9. Weinheim: Wiley-VCH; 2003. p. 63–80.
Simon R. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-mob transposon. Mol Gen Genom. 1984;196:413–20.
Desomer J, Dhaese P, Montagu MV. Transformation of Rhodococcus fascians by high-voltage electroporation and development of R. fascians cloning vectors. Appl Env Microbiol. 1990;56:2818–25.
Kalscheuer R, Arenskötter M, Steinbüchel A. Establishment of a gene transfer system for Rhodococcus opacus PD630 based on electroporation and its application for recombinant biosynthesis of poly(3-hydroxyalkanoic acids). Appl Microbiol Biotechnol. 1999;52:508–15.
Booth WT, Schlachter CR, Pote S, Ussin N, Mank NJ, Klapper V, Offermann LR, Tang C, Hurlburt BK, Chruszcz M. Impact of an N-terminal polyhistidine tag on protein thermal stability. ACS Omega. 2018;3:760–8.
Kimple ME, Brill AL, Pasker RL. Overview of affinity tags for protein purification. Curr Proto Protein Sci. 2013;73:1–26.
Rehm H, Letzel T. Der experimentator: proteinbiochemie/proteomics. Berlin: Springer-Verlag; 2016.
Wink M. Molekulare biotechnologie: Konzepte, Methoden und Anwendungen. Hoboken: Wiley; 2011. p. 1–654.
Paul CE, Eggerichs D, Westphal AH, Tischler D, Berke WJH. Flavoprotein monooxygenases: versatile biocatalysts. Biotechnol Adv. 2021;51:1–25.
Zhou Q, Peng S, Zhang K, Luo G, Han L, He Q, Tang. A Flavin-dependent monooxygenase mediates divergent oxidation of rifamycin. Org Lett. 2021;23:2342–6.
Pal S, Sengupta K. Computational-based insights into the phylogeny, structure, and function of Rhodococcus alkane-1-monooxygenase. 3 Biotech. 2020;10:391–7.
Zhu HJ, Zhang B, Wang L, Wang W, Liu SH, Igarashi Y, Bashiri G, Tan RX, Ge HM. Redox modifications in the biosynthesis of Alchivemycin A enable the formation of its key pharmacophore. J Am Chem Soc. 2021;143:4751–7.
Maier S, Heitzler T, Asmus K, Brçtz E, Hardter U, Hesselbach K, Paululat T, Bechthold A. Functional characterization of different ORFs including luciferase-like monooxygenase genes from the Mensacarcin gene cluster. Chem Biol Chem. 2015;16:1175–82.
Alali A, Zhang L, Li J, Zuo C, Wassouf D, Yan X, Schwarzer P, Günther S, Einsle O, Bechthold A. Biosynthesis of the tricyclic aromatic type II polyketide rishirilide: new potential third ring oxygenation after three cyclization steps. Mol Biotechnol. 2021;63:502–14.
Li S, Wen Y, Leng Y. Transcriptome analysis provides new insights into the tolerance and reduction of Lysinibacillus fusiformis 15–4 to hexavalent chromium. Appl Microbiol Biotechnol. 2021;105:7841–55.
Mascotti ML, Ayub MJ, Fraaije MW. On the diversity of F420-dependent oxidoreductases: a sequence- and structure-based classification. Proteins. 2021;89:1497–507.
Stecker C. Molekulare Charakterisierung des linearen Mega-Plasmides pBD2 aus Rhodococcus erythropolis BD2 und Identifizierung von Komponenten des Konjugationssystems. Cuvillier publishers 2004;1–244.
Crawford DL. Microbial transformations of low rank coals. Boca Raton: CRC Press; 1992. p. 1–240.
Etemadifar Z, Emtiazi G, Christofi N. Enhanced desulfurization activity in protoplast transformed Rhodococcus Erythropolis. Am-Eur J Agri Env Sci. 2008;3:285–91.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by Alexander von Humboldt (AvH) foundation, Germany (Ref No: IND 1162665 HFST-P). Dr. M. V. Reddy is grateful to the AvH foundation for providing the Postdoctoral fellowship.
Author information
Authors and Affiliations
Contributions
MVR and AS conceived and designed the research. MVR performed the experiments and analyzed the data. AS supervised the whole study. MVR wrote the original draft manuscript. AS reviewed and edited the manuscript. Both the authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all the authors, the corresponding authors state that there is no conflict of interest.
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Venkateswar Reddy, M., Steinbüchel, A. Evaluation of the function of a luciferase-like monooxygenase homologue in 4,4´-dithiodibutyric acid catabolism in Rhodococcus erythropolis MI2. Syst Microbiol and Biomanuf 2, 523–532 (2022). https://doi.org/10.1007/s43393-022-00080-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-022-00080-0