Abstract
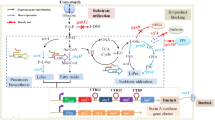
Lacto-N-neotetraose (LNnT), one of the most important human milk oligosaccharides, can be used as infants’ food additives. Nowadays, extraction, chemical and biological synthesis were utilized to obtain LNnT, while these methods still face some problems such as low yield and high cost. The aim of current work is to construct a de novo biosynthesis pathway of LNnT in E. coli K12 MG1655. The lgtA and lgtB were first expressed by a plasmid, resulting in a LNnT titer of 0.04 g/L. To improve the yield of LNnT on substrate lactose, lacZ and lacI were knocked out, and lacY was over-expressed. As a result, the yield of LNnT on lactose increased from 0.01 to 0.09 mol/mol, and the titer of LNnT elevated to 0.41 g/L. In addition, the pathway was regulated using the titer of Lacto-N-triose II (LNTII) as a measure, and obtained a high titer strain of LNnT for 1.04 g/L. Finally, the gene expressions were fine-tuned, the titer of LNnT reached 1.2 g/L, which was 93% higher than the control strain, and the yield on lactose reached 0.28 mol/mol. The engineering strategy of pathway construction and modulation used in this study is applicable to facilitate the microbial production of other metabolites in E. coli.





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
German JB, Freeman SL, Lebrilla CB, Mills DA. Human milk oligosaccharides: evolution, structures and bioselectivity as substrates for intestinal bacteria. In: Personalized nutrition for the diverse needs of infants and children; KARGER: Basel, 2008; pp. 205–22.
Duska-McEwen G, Senft AP, Ruetschilling TL, Barrett EG, Buck RH. Human milk oligosaccharides enhance innate immunity to respiratory syncytial virus and influenza in vitro. Food Nutr Sci. 2014;05:1387–98.
Victora CG, Bahl R, Barros AJD, França GVA, Horton S, Krasevec J, Murch S, Sankar MJ, Walker N, Rollins NC, et al. Breastfeeding in the 21st century: epidemiology, mechanisms, and lifelong effect. Lancet. 2016;387:475–90.
Chen R. The sweet branch of metabolic engineering: cherry-picking the low-hanging sugary fruits. Microb Cell Fact. 2015;14:1–10.
Sprenger N, Lee LY, De Castro CA, Steenhout P, Thakkar SK. Longitudinal change of selected human milk oligosaccharides and association to infants’ growth, an observatory, single center, longitudinal cohort study. PLoS ONE. 2017;12:1–15.
Coppa GV, Pierani P, Zampini L, Bruni S, Carloni I, Gabrielli O. Characterization of oligosaccharides in milk and feces of breast-fed infants by high-performance anion-exchange chromatography. Adv Exp Med Biol. 2001;501:307–14.
Petschacher B, Nidetzky B. Biotechnological production of fucosylated human milk oligosaccharides: Prokaryotic fucosyltransferases and their use in biocatalytic cascades or whole cell conversion systems. J Biotechnol. 2016;235:61–83.
Vandenplas Y, Berger B, Carnielli VP, Ksiazyk J, Lagstrom H, Luna SM, Migacheva N, Mosselmans J-M, Picaud J-C, Possner M, et al. Human milk oligosaccharides: 2-fucosyllactose (2-FL) and lacto-N-neotetraose (LNnT) in infant formula. Nutrients. 2018;10:1161.
Elison E, Vigsnaes LK, Krogsgaard RL, Rasmussen J, Sorensen N, McConnell B, Hennet T, Sommer MOA, Bytzer P. Oral supplementation of healthy adults with 2′-O-fucosyllactose and lacto-N-neotetraose is well tolerated and shifts the intestinal microbiota. Br J Nutr. 2016;116:1356–68.
Huang D, Yang K, Liu J, Xu Y, Wang Y, Wang R, Liu B, Feng L. Metabolic engineering of Escherichia coli for the production of 2′-fucosyllactose and 3-fucosyllactose through modular pathway enhancement. Metab Eng. 2017;41:23–38.
Hollands K, Baron CM, Gibson KJ, Kelly KJ, Krasley EA, Laffend LA, Lauchli RM, Maggio-Hall LA, Nelson MJ, Prasad JC, et al. Engineering two species of yeast as cell factories for 2′-fucosyllactose. Metab Eng. 2019;52:232–42.
Soo N, Kim T, Park Y-C, Kim J, Seo J-H, Han NS, Kim T, Park Y-C, Kim J, Seo J-H. Biotechnological production of human milk oligosaccharides. Biotechnol Adv. 2012;30:1268–78.
Smilowitz JT, Lebrilla CB, Mills DA, German JB, Freeman SL. Breast milk oligosaccharides: Structure-function relationships in the neonate. Annu Rev Nutr. 2014;34:143–69.
Priem B, Gilbert M, Wakarchuk WW, Heyraud A, Samain E. A new fermentation process allows large-scale production of human milk oligosaccharides by metabolically engineered bacteria. Glycobiology. 2002;12:235–40.
Bode L. Recent advances on structure, metabolism, and function of human milk oligosaccharides. J Nutr. 2006;136:2127–30.
Sprenger GA, Baumgärtner F, Albermann C. Production of human milk oligosaccharides by enzymatic and whole-cell microbial biotransformations. J Biotechnol. 2017;258:79–91.
Jin W, Zhang F, Linhardt RJ. Bioengineered production of glycosaminoglycans and their analogues. Syst Microbiol Biomanuf. 2020;1:1–8.
Deng J, Lv X, Li J, Du G, Chen J, Liu L. Recent advances and challenges in microbial production of human milk oligosaccharides. Syst Microbiol Biomanufacturing. 2020;1:3.
Bych K, Mikš MH, Johanson T, Hederos MJ, Vigsnæs LK, Becker P. Production of HMOs using microbial hosts—from cell engineering to large scale production. Curr Opin Biotechnol. 2019;56:130–7.
Jennings MP, Hood DW, Peak LRA, Virji M, Moxon ER. Molecular analysis of a locus for the biosynthesis and phase-variable expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. Mol Microbiol. 1995;18:729–40.
Heyraud A, Priem B, Dumon C, Bosso C, Martin LS. In vivo fucosylation of lacto-N-neotetraose and lacto-N-neohexaose by heterologous expression of Helicobacter pylori a-1,3 fucosyltransferase in engineered Escherichia coli. Glycoconj J. 2001;6:465–474.
Dong X, Li N, Liu Z, Lv X, Li J, Du G, Wang M, Liu L. Modular pathway engineering of key precursor supply pathways for lacto-N-neotetraose production in Bacillus subtilis. Biotechnol Biofuels. 2019;12:1–11.
Dong X, Li N, Liu Z, Lv X, Shen Y, Li J, Du G, Wang M, Liu L. CRISPRi-guided multiplexed fine-tuning of metabolic flux for enhanced lacto-N-neotetraose production in Bacillus subtilis. J Agric Food Chem. 2020;68:2477–84.
Baumgartner F, Conrad J, Sprenger GA, Albermann C. Synthesis of the human milk oligosaccharide lacto-N-tetraose in metabolically engineered, plasmid-free E. coli. ChemBioChem. 2014;15:1896–900.
Jung SM, Park YC, Seo JH. Production of 3-fucosyllactose in engineered Escherichia coli with α-1,3-fucosyltransferase from Helicobacter pylori. Biotechnol J. 2019;14:1–7.
Wood EJ. Molecular cloning. A laboratory manual by T Maniatis, E F Fritsch and J Sambrook. New York: Cold Spring Harbor Laboratory; 1982. p. 545.
Chung ME, Yeh IH, Sung LY, Wu MY, Chao YP, Ng IS, Hu YC. Enhanced integration of large DNA into E. coli chromosome by CRISPR/Cas9. Biotechnol Bioeng. 2017;114:172–83.
Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81:2506–14.
Englaender JA, Jones JA, Cress BF, Kuhlman TE, Linhardt RJ, Koffas MAG. Effect of genomic integration location on heterologous protein expression and metabolic engineering in E. coli. ACS Synth Biol. 2017;6:710–20.
Juhas M, Evans LDB, Frost J, Davenport PW, Yarkoni O, Fraser GM, Ajioka JW. Escherichia coli flagellar genes as target sites for integration and expression of genetic circuits. PLoS ONE. 2014;9:1–7.
Sabri S, Steen JA, Bongers M, Nielsen LK, Vickers CE. Knock-in/Knock-out (KIKO) vectors for rapid integration of large DNA sequences, including whole metabolic pathways, onto the Escherichia coli chromosome at well-characterised loci. Microb Cell Fact. 2013;12:1–15.
Juhas M, Ajioka JW. Lambda Red recombinase-mediated integration of the high molecular weight DNA into the Escherichia coli chromosome. Microb Cell Fact. 2016;15:1–10.
Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–96.
Gu L, Yuan H, Lv X, Li G, Cong R, Li J, Du G, Liu L. High-yield and plasmid-free biocatalytic production of 5-methylpyrazine-2-carboxylic acid by combinatorial genetic elements engineering and genome engineering of Escherichia coli. Enzyme Microb Technol. 2020;134:109488.
Hossain GS, Li J, Shin HD, Chen RR, Du G, Liu L, Chen J. Bioconversion of l-glutamic acid to α-ketoglutaric acid by an immobilized whole-cell biocatalyst expressing l-amino acid deaminase from Proteus mirabilis. J Biotechnol. 2014;169:112–20.
Albermann C, Piepersberg W, Wehmeier UF. Synthesis of the milk oligosaccharide 2′-fucosyllactose using recombinant bacterial enzymes. Carbohydr Res. 2001;334:97–103.
Chin YW, Kim JY, Lee WH, Seo JH. Enhanced production of 2’-fucosyllactose in engineered Escherichia coli BL21star(DE3) by modulation of lactose metabolism and fucosyltransferase. J Biotechnol. 2015;210:107–15.
Baumgärtner F, Jurzitza L, Conrad J, Beifuss U, Sprenger GA, Albermann C. Synthesis of fucosylated lacto-N-tetraose using whole-cell biotransformation. Bioorganic Med Chem. 2015;23:6799–806.
Funding
This work was supported by the National Natural Science Foundation of China (31930085, 32021005), and the key research and development program of China (2018YFA0900300, 2020YFA0908300).
Author information
Authors and Affiliations
Contributions
WZ has a major contribution to the study, and writing of the manuscript. ZML, GMY, NL and XQL analyzed the experimental data. XMD, LYF, LJH and DGC revised the manuscript. LL conceived and supervised the project and contributed to the writing of the manuscript. All the authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, W., Liu, Z., Gong, M. et al. Metabolic engineering of Escherichia coli for the production of Lacto-N-neotetraose (LNnT). Syst Microbiol and Biomanuf 1, 291–301 (2021). https://doi.org/10.1007/s43393-021-00023-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-021-00023-1