Abstract
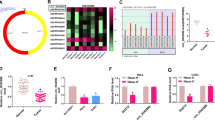
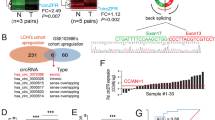
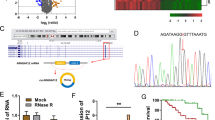
Cervical cancer (CC) is the most serious gynecological malignancy among women worldwide. As a subtype of noncoding RNAs (ncRNAs), circular RNAs (circRNAs) play important roles in the regulation of gene expression and cancer progression. It was discovered from the cancer-specific circRNA database (CSCD) that circ_0019435 was mainly distributed in the nucleus of HeLa-S3 cells. However, few researches have mentioned circ_0019435 with its function in cancers. The present study uncovered that circ_0019435 was upregulated in CC cells by qRT-PCR. Moreover, circ_0019435 was more stable than its linear isoform-ABCC2. Besides, no regulation of circ_0019435 on ABCC2 and the chemoresistance of CC cells were found. Then, it was unveiled by a series of functional assays including colony formation, trypan blue staining, and transwell invasion assays in that circ_0019435 ablation induced the suppression of proliferation, invasion, and EMT of HeLa and SiHa cells. The subcellular distribution of circ_0019435 was assessed by subcellular fractionation and FISH assay. Furthermore, it was disclosed that circ_0019435 binds to EZH2 to silence DKK1 and PTEN. Finally, rescue assays corroborated that DKK1 and PTEN were involved in circ_0019435-mediated CC cell progression. In conclusion, circ_0019435 regulates DKK1 and PTEN expression at the epigenetic level, thereby influencing the progression of CC cells.




Similar content being viewed by others
Data Availability
Not applicable.
References
Siva S, Deb S, Young RJ, Hicks RJ, Callahan J, Bressel M, et al. (1)(8)F-FDG PET/CT following chemoradiation of uterine cervix cancer provides powerful prognostic stratification independent of HPV status: a prospective cohort of 105 women with mature survival data. Eur J Nucl Med Mol Imaging. 2015;42(12):1825–32. https://doi.org/10.1007/s00259-015-3112-8.
Sherman SM, Lane EL. Awareness of risk factors for breast, lung and cervical cancer in a UK student population. Journal of cancer education : the official journal of the American Association for Cancer Education. 2015;30(4):660–3. https://doi.org/10.1007/s13187-014-0770-3.
Zhang HH, Li AH. Long non-coding RNA FEZF1-AS1 is up-regulated and associated with poor prognosis in patients with cervical cancer. Eur Rev Med Pharmacol Sci. 2018;22(11):3357–62. https://doi.org/10.26355/eurrev_201806_15156.
Sakuragi N. Refining insight into cervical cancer progression. Lancet Oncol. 2014;15(4):371–2. https://doi.org/10.1016/s1470-2045(14)70085-3.
Casarin J, Bogani G, Serati M, Pinelli C, Laganà AS, Garzon S, et al. Presence of glandular cells at the preoperative cervical cytology and local recurrence in endometrial cancer. Int J Gynecol Pathol. 2020;39(6):522–8. https://doi.org/10.1097/pgp.0000000000000642.
Casarin J, Bogani G, Papadia A, Ditto A, Pinelli C, Garzon S, et al. Preoperative conization and risk of recurrence in patients undergoing laparoscopic radical hysterectomy for early stage cervical cancer: a multicenter study. J Minim Invasive Gynecol. 2021;28(1):117–23. https://doi.org/10.1016/j.jmig.2020.04.015.
Valenti G, Vitale SG, Tropea A, Biondi A, Laganà AS. Tumor markers of uterine cervical cancer: a new scenario to guide surgical practice? Updat Surg. 2017;69(4):441–9. https://doi.org/10.1007/s13304-017-0491-3.
Rossetti D, Vitale SG, Tropea A, Biondi A, Laganà AS. New procedures for the identification of sentinel lymph node: shaping the horizon of future management in early stage uterine cervical cancer. Updat Surg. 2017;69(3):383–8. https://doi.org/10.1007/s13304-017-0456-6.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Smolle E, Haybaeck J. Non-coding RNAs and lipid metabolism. Int J Mol Sci. 2014;15(8):13494–513. https://doi.org/10.3390/ijms150813494.
Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–8. https://doi.org/10.1080/15476286.2015.1020271.
Zhou ZB, Huang GX, Fu Q, Han B, Lu JJ, Chen AM, et al. circRNA.33186 contributes to the pathogenesis of osteoarthritis by sponging miR-127-5p. Mol Ther. 2019;27(3):531–41. https://doi.org/10.1016/j.ymthe.2019.01.006.
Liu J, Xue N, Guo Y, Niu K, Gao L, Zhang S, et al. CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging. 2019;11(24):12412–27. https://doi.org/10.18632/aging.102580.
Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. https://doi.org/10.1186/s13059-014-0571-3.
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. https://doi.org/10.1186/s12943-017-0663-2.
Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J Neurosci Res. 2018. https://doi.org/10.1002/jnr.24356.
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–58. https://doi.org/10.1093/nar/gkw027.
Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein-lncRNA interaction. Brief Bioinform. 2016;17(1):106–16. https://doi.org/10.1093/bib/bbv031.
Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics, proteomics & bioinformatics. 2017;15(3):177–86. https://doi.org/10.1016/j.gpb.2016.12.005.
Ding J, Li J, Wang H, Tian Y, Xie M, He X, et al. Long noncoding RNA CRNDE promotes colorectal cancer cell proliferation via epigenetically silencing DUSP5/CDKN1A expression. Cell Death Dis. 2017;8(8):e2997. https://doi.org/10.1038/cddis.2017.328.
Zhang E, He X, Yin D, Han L, Qiu M, Xu T, et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109. https://doi.org/10.1038/cddis.2015.356.
Comsa E, Nguyen KA, Loghin F, Boumendjel A, Peuchmaur M, Andrieu T, et al. Ovarian cancer cells cisplatin sensitization agents selected by mass cytometry target ABCC2 inhibition. Future Med Chem. 2018;10(11):1349–60. https://doi.org/10.4155/fmc-2017-0308.
Guo J, Liu M, Yang D, Bouxsein ML, Saito H, Galvin RJ, et al. Suppression of Wnt signaling by Dkk1 attenuates PTH-mediated stromal cell response and new bone formation. Cell Metab. 2010;11(2):161–71. https://doi.org/10.1016/j.cmet.2009.12.007.
Benhamou D, Labi V, Getahun A, Benchetrit E, Dowery R, Rajewsky K, et al. The c-Myc/miR17-92/PTEN axis tunes PI3K activity to control expression of recombination activating genes in early B cell development. Front Immunol. 2018;9:2715. https://doi.org/10.3389/fimmu.2018.02715.
Song T, Xu A, Zhang Z, Gao F, Zhao L, Chen X, et al. CircRNA hsa_circRNA_101996 increases cervical cancer proliferation and invasion through activating TPX2 expression by restraining miR-8075. J Cell Physiol. 2019;234(8):14296–305. https://doi.org/10.1002/jcp.28128.
Liu J, Wang D, Long Z, Liu J, Li W. CircRNA8924 promotes cervical cancer cell proliferation, migration and invasion by competitively binding to MiR-518d-5p /519-5p family and modulating the expression of CBX8. Cell Physiol Biochem. 2018;48(1):173–84. https://doi.org/10.1159/000491716.
Cai H, Zhang P, Xu M, Yan L, Liu N, Wu X. Circular RNA hsa_circ_0000263 participates in cervical cancer development by regulating target gene of miR-150-5p. J Cell Physiol. 2019;234(7):11391–400. https://doi.org/10.1002/jcp.27796.
Shen CJ, Cheng YM, Wang CL. LncRNA PVT1 epigenetically silences miR-195 and modulates EMT and chemoresistance in cervical cancer cells. J Drug Target. 2017;25(7):637–44. https://doi.org/10.1080/1061186x.2017.1307379.
Su M, Xiao Y, Tang J, Wu J, Ma J, Tian B, et al. Role of lncRNA and EZH2 interaction/regulatory network in lung cancer. J Cancer. 2018;9(22):4156–65. https://doi.org/10.7150/jca.27098.
Ling Z, Wang X, Tao T, Zhang L, Guan H, You Z, et al. Involvement of aberrantly activated HOTAIR/EZH2/miR-193a feedback loop in progression of prostate cancer. J Exp Clin Cancer Res. 2017;36(1):159. https://doi.org/10.1186/s13046-017-0629-7.
Acknowledgements
We appreciate all the participants who provide supports for this research.
IRB Approval
Not applicable.
Funding
This study was supported by Ningbo Natural Science Foundation (Grant No. 2019A610305) and Medical Scientific Research Foundation of Zhejiang Province (Grant No. 2020KY832).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare that no competing interest exists in this study.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Wang, Q., Zhuo, Z., Yu, H. et al. Circ_0019435 Exerts Its Functions in the Cellular Process of Cervical Cancer via Epigenetically Silencing DKK1 and PTEN. Reprod. Sci. 28, 2989–2999 (2021). https://doi.org/10.1007/s43032-021-00625-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-021-00625-z