Abstract
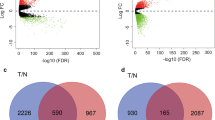
Improved insight into the molecular mechanisms of triple-negative breast cancer (TNBC) is required to predict prognosis and develop a new therapeutic strategy for targeted genes. The aim of this study was to identify genes significantly associated with TNBC and further analyze their prognostic significance. The Cancer Genome Atlas (TCGA) TNBC database and gene expression profiles of GSE76275 from Gene Expression Omnibus (GEO) were used to explore differentially co-expressed genes in TNBC compared with those in normal tissues and non-TNBC breast cancer tissues. Differential gene expression and weighted gene co-expression network analyses identified 24 differentially co-expressed genes. Functional annotation suggested that these genes were primarily enriched in processes such as metabolism, membrane, and protein binding. The protein-protein interaction (PPI) network further identified ten hub genes, five of which (MAPT, CBS, SOX11, IL6ST, and MEX3A) were confirmed to be differentially expressed in an independent dataset (GSE38959). Moreover, CBS and MEX3A expression was upregulated, whereas IL6ST expression was downregulated in TNBC tissues compared to that in other breast cancer subtypes. Furthermore, lower expression of IL6ST was associated with worse overall survival in patients with TNBC. Thus, IL6ST might play an important role in TNBC progression and could serve as a tumor suppressor gene for diagnosis and treatment.











Similar content being viewed by others
Data Availability
All datasets analyzed in this study can be found at TCGA (https://portal.gdc.cancer.gov/), GEO (https://www.ncbi.nlm.nih.gov/gds), and Oncomine (https://www.oncomine.org/resource/login.html).
Code Availability
Not applicable.
References
Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:194153.
Siegel RL, Miller KD, Sauer GA, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:14564.
Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–74.
Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Weigelt B, Horlings H, Kreike B, Hayes M, Hauptmann M, Wessels L, et al. Refinement of breast cancer classification by molecular characterization of histological special types. J Pathol. 2008;216:141–50.
Liao Y, Liao Y, Li J, Li J, Fan Y, Xu B. Polymorphisms in AURKA and AURKB are associated with the survival of triple-negative breast cancer patients treated with taxane-based adjuvant chemotherapy. Cancer Manag Res. 2018;10:3801–8.
Vagia E, Mahalingam D, Cristofanilli M. The landscape of targeted therapies in TNBC. Cancers (Basel). 2020;12:916.
Huang D, Huang Y, Huang Z, Weng J, Zhang S, Gu W. Relation of AURKB over expression to low survival rate in BCRA and reversine modulated aurora B kinase in breast cancer cell lines. Cancer Cell Int. 2019;19:166.
Karra H, Repo H, Ahonen I, Löyttyniemi E, Pitkänen R, Lintunen M, et al. Cdc20 and securin overexpression predict short-term breast cancer survival. Br J Cancer. 2014;110:2905–13.
Paul D, Ghorai S, Dinesh US, Shetty P, Chattopadhyay S, Santra MK. Cdc20 directs proteasome-mediated degradation of the tumor suppressor SMAR1 in higher grades of cancer through the anaphase promoting complex. Cell Death Dis. 2017;8:e2882.
Deng JL, Xu YH, Wang G. Identification of potential crucial genes and key pathways in breast cancer using bioinformatic analysis. Front Genet. 2019;10:695.
Zhang WL, Wang WN, Sun YX, Bi LQ. Screening crucial genes involved in triple-negative breast cancer through bioinformatics analysis of microarray data. Eur J Gynaecol Oncol. 2018;39:101–7.
Jiang YQ, Liu Y, Tan X, Yu S, Luo J. TPX2 as a novel prognostic indicator and promising therapeutic target in triple-negative breast cancer. Clin Breast Cancer. 2019;19:450–5.
Chen J, Qian X, He Y, Han X, Pan Y. Novel key genes in triple-negative breast cancer identified by weighted gene co-expression network analysis. J Cell Biochem. 2019;120:16900–12.
Naorem LD, Muthaiyan M, Venkatesan A. Integrated network analysis and machine learning approach for the identification of key genes of triple-negative breast cancer. J Cell Biochem. 2019;120:6154–67.
Wang YJ, Kojetin D, Burris TP. Anti-proliferative actions of a synthetic REV-ERB alpha/beta agonist in breast cancer cells. Biochem Pharmacol. 2015;96:315–22.
Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559.
Segundo-Val IS, Sanz-Lozano CS. Introduction to the gene expression analysis. Methods Mol Biol. 2016;1434:29–43.
Li CY, Cai JH, Tsai JJP, Wang CCN. Identification of hub genes associated with development of head and neck squamous cell carcinoma by integrated bioinformatics analysis. Front Oncol. 2020;10:681.
Wu X, Zou T, Cao N, Ni J, Xu W, Zhou T, et al. Plasma homocysteine levels and genetic polymorphisms in folate metabolism are associated with breast cancer risk in Chinese women. Hered Cancer Clin Pract. 2014;12:2.
Kawahara B, Moller T, Hu-Moore K, Carrington S, Faull KF, Sen S, et al. Attenuation of antioxidant capacity in human breast cancer cells by carbon monoxide through inhibition of cystathionine β-synthase activity: implications in chemotherapeutic drug sensitivity. J Med Chem. 2017;60:8000–10.
Jiang H, Zhang X, Luo J, Dong C, Xue J, Wei W, et al. Knockdown of hMex-3A by small RNA interference suppresses cell proliferation and migration in human gastric cancer cells. Mol Med Rep. 2012;6:575–80.
Krepischi ACV, Maschietto M, Ferreira EN, Silva AG, Costa SS, da Cunha IW, et al. Genomic imbalances pinpoint potential oncogenes and tumor suppressors in Wilms tumors. Mol Cytogenet. 2016;9:20.
Yang D, Jiao Y, Li Y, Fang X. Clinical characteristics and prognostic value of MEX3A mRNA in liver cancer. PeerJ. 2020;8:e8252.
Wang X, Shan YQ, Tan QQ, Tan CL, Zhang H, Liu JH, et al. MEX3A knockdown inhibits the development of pancreatic ductal adenocarcinoma. Cancer Cell Int. 2020;20:63.
Funding
This study was supported by the Fund of Youth Innovation of Inner Mongolia Medical University [grant number YKD2020QNCX021].
Author information
Authors and Affiliations
Contributions
Conceptualization, R.J. and YJ.W.; methodology, ZX.L.; software, W.L.; formal analysis, YC.J.; resources, Y.L.; writing—original draft preparation, R.J.; writing—review and editing, PF.N. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Ethical Consents
This study is conducted based on TCGA and GEO databases and is not involved in animal or human experiments, so ethical consents are not appropriate in this study.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jia, R., Weng, Y., Li, Z. et al. Bioinformatics Analysis Identifies IL6ST as a Potential Tumor Suppressor Gene for Triple-Negative Breast Cancer. Reprod. Sci. 28, 2331–2341 (2021). https://doi.org/10.1007/s43032-021-00509-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-021-00509-2