Abstract
Autophagy is a conserved, lysosome-dependent catabolic process of eukaryotic cells which is involved in cellular differentiation. Here, we studied its specific role in the differentiation of spermatogonial cells in the Drosophila testis. In the apical part of the Drosophila testis, there is a niche of germline stem cells (GSCs), which are connected to hub cells. Hub cells emit a ligand for bone morhphogenetic protein (BMP)-mediated signalling that represses Bam (bag of marbles) expression in GSCs to maintain them in an undifferentiated state. GSCs divide asymmetrically, and one of the daughter cells differentiates into a gonialblast, which eventually generates a cluster of spermatogonia (SG) by mitoses. Bam is active in SG, and defects in Bam function arrest these cells at mitosis. We show that BMP signalling represses autophagy in GSCs, but upregulates the process in SG. Inhibiting autophagy in SG results in an overproliferating phenotype similar to that caused by bam mutations. Furthermore, Bam deficiency leads to a failure in downstream mechanisms of the autophagic breakdown. These results suggest that the BMP-Bam signalling axis regulates developmental autophagy in the Drosophila testis, and that acidic breakdown of cellular materials is required for spermatogonial differentiation.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
Introduction
Autophagy, also known as cellular self-digestion, is a highly conserved catabolic process of eukaryotic cells. Through degrading damaged and superfluous cellular materials (organelles and macromolecules), the autophagic process plays a fundamental role in cellular homeostasis and macromolecule turnover. A specific type of autophagy is called macroautophagy (hereafter is referred to as autophagy) that is initiated by growing of a double-membraned structure. When this membrane formation becomes completed, a vesicule called autophagosome is formed, which contains the sequestered cytoplasmic materials destined for degradation. The autophagosome then fuses with a lysosome to form an autolysosome, in which the enzymatic breakdown takes place by acidic hydrolases. Autophagy serves as a dynamic recycling system that provides building blocks and energy for cellular renovation and homeostasis (Parzych and Klionsky 2014).
Autophagy fulfills multiple roles in various cellular processes, such as maintaining homeostasis, energy production, and differentiation (Mizushima and Levine 2010). Autophagy has a significant importance in the differentiation of germline stem cells (GSCs) as it maintains a balance between self-renewal and differentiation. Defects in this balance can lead to tumorigenesis and tissue hyperplasia, as well as tissue degeneration and cellular aging (Cheng et al. 2011).
The early stage of spermatogenesis in the testis of the fruitfly Drosophila melanogaster represents a well-establsihed model for studying the role of autophagy in differentiation (de Cuevas and Mautnis 2011). There are two types of stem cells residing in the fly testis, germline stem cells (GSCs) and cyst stem cells (CySCs). These two specific stem cells are physically connected with a non-dividing stromal cell group, called hub. The hub serves as a niche to provide an adequate microenvironment for stems cells. Hub cells supply important signals and cellular junction for these cells. Through distinct steps of asymmetric division of GSCs, one daughter cell preserves its stem cell identity, while the other differentiates into a gonialblast, which then forms spermatogonia (SG) through a series of mitosis (de Cuevas and Mautnis 2011). During the early stage of spermatogenesis, germ cells go through two cell fate changes. The first can be observed during the asymmetric division of GSCs, when the two daughter cells choose different cell fates. The second one takes place when SG enter into the meiotic stage (Fig. 1 A). There is a key importance in transmitting signaling ligands originating from the hub toward stem cells, because these factors maintain stem cells in an undifferentiated state. These ligands activate signaling pathways such as JAK/STAT (Janus kinase/signal transducer and activator of transcription proteins) and BMP (bone morphogenetic protein). Defects in these signalling pathways often lead to a premature differentiation of stem cells (Dansereau and Lasko 2008).
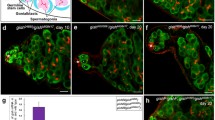
A Early Spermatogenesis of Drosophila: Two types of stem cell populations take part in spermatogenesis, which are located at the proximal site of the Drosophila testis in the germ chamber, also known as the germarium. They are anchored to a unique non-dividing (post-mitotic) somatic cell population called hub. The stem cell populations consist of germ stem cells (GSCs) and cyst stem cells (CySCs). Asymmetric division of GSCs produces two daughter cells with different characteristics, one of them remains stem cell while the other differentiates further. This later one is called gonialblast (Gb), which goes through four mitotic divisions to produce a cluster of 16 spermatogonial cells (SG), which are connected to each other by cytoplasmic bridges. Cells of the cluster of 16 spermatogonial cells are called spermatocytes, which enter into the meiotic phase, in which they differentiate into sperms through several steps. Asymmetric division of the other stem cell population, CySCs, provides a differentiating daughter cell termed as the cyst cell (CC), two of which envelops a Gb, and later on during spermatogenesis, it surrounds the spermatogonial cluster by continuing to grow. B The BMP (Dpp/Gbb) signaling pathway is responsible for maintaining GSCs and preserving them in an undifferentiated state. BMP proteins belong to the TGFβ (Transforming Growth Factor β) superfamily. Two distinct BMP ligands can be found in the germline of Drosophila, these are Dpp (decapentaplegic) and Gbb (glass bottom boat). Both of these ligands activate heterodimer receptors by forming homo- or heterodimers. Heterodimer receptors contain a Type I protein such as Sax (Saxophone) or Tkv (Thickveins), and the other component is Type II protein, like Pnt (Punt). Receptor activation leads to the phosphorylation of one of the citoplasmic remote control, for instance Mad (Mothers against dpp, ortholog of the mammalian co-Smads) becomes pMad. pMad then forms with Med (Medea, the ortholog of mammalian R-Smads) protein either a heterodimer or a heterotrimer in 2:1 ratio (Mad:Med), resulting in expressional changes of target genes. Among several mechanisms, this effect occurs by the repression of translation of the Bam differentiation-promotion factor. C and C’: The mechanism of the Bam complex and the expressional distribution of the members of the complex in early spermatogenesis. Activation of the BMP pathway in GSCs (also in the ovary of Drosophila) leads to inhibition of the bam differentiation factor gene. As a result, Bam is inactive in GSCs, while in daughter cells, it differentiates into SG, where the BMP pathway is inactive and Bam is active. Its expression increases progressively in SG. Upon Bam loss, the meiotic phase will not occur, SG remain in mitosis, resulting in larger cysts. This phenotype is known as bag of marbles. C’ Bam forms a complex with Bgcn and Tut. C) Expression of Tut is increased at the level of differentiation and displays a peak in the cluster of 16-cell-SG, while the expression of Bgcn remains constant during early spermatogenesis. C’ The complex of Tut-Bam-Bgcn inhibits the expression of target genes, such as mei-P26, by binding 3’UTR of their mRNA
BMP signaling functions in maintaining a balance between differentiation and self-renewal of the stem cell niche. In Drosophila, this pathway has three different ligands, which are encoded by the dpp (decapentaplegic), gbb (glass bottom boat) and scw (screw) genes (Bangi and Wharton 2006b). The ligands form a homo- or heterodimer, then activate the transmission of the signal through heterodimer receptors such as Type I Thickveins (Tkv) or Type II Punt (Pnt). Activated thickveins in turn phosphorilates Mad, a citoplasmic remote control, which leads to pMad formation. PMad forming a complex with Mad accumulates in the nucleus, modulating the expression of target genes (e.g., bam) through binding distinct sequence motifs (Hamaratoglu et al. 2014). In the case of regulating bam transcription, transmission of the signal results in changes in stem cell fates. Signals determining stem cell fates can reach only those cells that establish a physical contact with hub (thus BMP signalling affects cells that are adjacent to hub). In the Drosophila testis, the dosage-dependent concentration and spatial distribution of BMP signaling have a critical role in preserving stem cell identity (Hayashi et al. 2009). BMP signaling can be modulated at several levels, including the expression and transmission of the ligands and receptors, through extracellular modulators, components of the extracellular matrix, post-translational modifications of the Mad/Med complex, and the target genes they regulate (Fig. 1 B) (Araujo et al. 2011). Modulating the activity of target genes can be performed by distinct mechanisms of derepression, silencing and direct activation. BMP signalling also shows a great importance in the self-renewal of GSCs. Specific BMP ligands, especially Dpp and Gbb, are secreted by both hub and CySCs. Thus, signal transmission can be activated only in GSCs (De Cuevas and Matunis 2011). BMP signalling emitted by the hub represses the differentiation factor Bam (bag of marbles) in GSCs. In SG, where BMP is switched off, Bam is activated (Fig. 1 B) (De Cuevas and Matunis 2011). Bam forms a complex with Bgcn (Benign gonial cell neoplasm) and Tut (Tumorous testis). The complex initiatiates the proliferation of SG and promotes their differentiation (Chen et al. 2014). It has been shown that the cell fate is determined by a critical treshold of Bam concentration (Insco et al. 2009, 2012). Bam and Tut levels reach the highest peak right before the last meiotic division, when they promote the activation of a spermatocyte differentiation program (Fig. 1 C). In preserving the stemness fate, there is an inhibitory signal that serves as an obstacle to prevent cells from undergoing differentiation. The level of the inhibitory signal decreases as Bam concentration becomes elevated. The accumulation of Bam in SG is stimulated by a TRIM-NHL (TRIpartite Motif-NHL) tumor suppressor protein, Mei-P26, which is required for the differentiation of the GSC lineage (Insco et al. 2012). Tut binds preferentially to a long isoform of Mei-P26 3′UTR and is essential for the translational repression of a Mei-P26 reporter. Both Bam and Bgcn are RBPs (RNAi-binding proteins) that repress mei-P26 expression. A genetic analysis revealed that Tut, Bam and Bgcn are each required for repressing mei-P26 and stimulating GSC differentiation. These proteins can be anchored to each other to form a complex, in which Bam holds Tut on the N-terminus and Bgcn on the C-terminus (Chen et al. 2014). Bam and Bgcn together form a negative feedback loop by connecting to the 3’UTR of Mei-P26, thereby inhibiting its translation. However, little is known about the mechanism by which the complex binds to mRNA (Li et al. 2013).
In the Drosophila ovary and testis, activation of BMP signaling in the GSCs results in the suppression of the Bam differentiation-promoting factor. bam downregulation cannot be transmitted to the daughter cell that undergoes differentiation into SG. Bam activity progressively increases as SG proliferate.
In the absence of Bam activity, SG cannot enter into the meiotic stage, resulting in overproliferation of unmature germ cells being stucked in the mitotic phase. In this case, there is a distinct morphological characteristics of the testis which is known as the bag-of-marbles phenotype (Gönczy and DiNardo 1997). Ectopic expression of bam leads to the absence of GSCs in the niche. In this study, we asked whether and how autophagy is involved in SG differenciation.
Materials and Methods
Fly stocks and their maintenance
Flies were kept at 25 °C or 32 °C (in the case of bam overexpression) on normal fly cornmeal nutrient (Billes et al.2018). For maintenance of fruit fly strains and characterisations of the vital. Strains were ordered from Bloomington Drosophila Stock Center (BDSC) or kindly provided by other researchers or generated and described by us earlier.
Isogenic w1118 (BDSC: 5905), Atg1-TRiP (BDSC: 35,177, Kuhn et al., 2015), Atg2 -TRiP (BDSC: 34,719, Bonfini et al. 2021), Atg5-TRiP (BDSC: 34,899, Kuhn et al., 2015, Atg14-TRiP (BDSC:40,858,Cunningham et al., 2020), Atg16-TRiP (BDSC: 34,358, Palmer et al., 2020), Atg7Δ77, Atg7Δ14, Aut1/Atg3 10 (provided by Gábor Juhász-Department of Anatomy, Cell- and Developmental Biology, Eötvös Loránd University, Budapest, Hungary, Juhász et al. 2003), hs-bam (BDSC: 24,636, Lim et al., 2015), bam-TRiP (BDSC: 33,631, Teixeira et al., 2015), bamΔ86 (BDSC: 5427, McKearin and Ohlstein 1995), Df (3R)FFD-0089346/TM6C (BDSC: 27,402), tut -TRiP (BDSC: 36,752), bgcn-TRiP (BDSC: 36,636), mad -TRiP (BDSC: 35,648, Follansbee et al., 2017), tkv4 {neoFRT}40A (BDSC: 51,297), P{Ubi-GFP.D}33 P{Ubi-GFP.D}38 P{neoFRT}40A (BDSC: 5189, Li et al., 2019),P{hsFLP}12, y1 w*; tkv4 P{neoFRT}40A/CyO(BDSC: 51,297) and P{hsFLP}12, w*; FRT40A (provided by Gábor Juhász-Department of Anatomy, Cell- and Developmental Biology, Eötvös Loránd University, Budapest, Hungary), tkvQ253D.Nb (BDSC: 36,536, Blatt et al., 2021), UAS-dpp (BDSC: 1486, Follansbee et al., 2017), UAS-myr-GFP (BDSC: 32,199, Kondo et al., 2020). Bam-Gal4, bab-Gal4 and Nanos-Gal4 (provided by Rita Sinka-Department of Genetics, University of Szeged, Szeged, Hungary).
Vital dyes
LysoTracker Red (Life Technologies, L-7528, diluted in 1:1000 ratio in PBS) was applied for staining acidic compartments. Nuclei were stained with 50 μg Hoechst in glycerol/PBS (4:1) cover solution. Adult testes were pre-dissected in PBS, then 65 µl LysoTR dye was pipetted into the samples. Samples were incubated in LysoTR for 2.5 min in dark, then the dye was removed by rinsing with 100 µl PBS, washing with PBS for another two times for 2 min, after the last wash, testes were dissected entirely. 13,5 µl cover solution was applied.
Drug treatment
Flies were maintained on standard cornmeal-sugar agar medium. Animals were placed into vials containing treated medium (heat-treated yeast suspension) immediately after eclosion for a day. Bafilomycin dissolved in DMSO (Sigma, B1793, final concentration was 50 nM), and Wortmannin was used in 50 µM final volume (Sigma, W1628, diluted in DMSO). For control, the same volume DMSO without Bafilomycin and Wortmannin was used. Rapamycin (Sigma, R0395) was made at a concentration of 50 mM in 100% EtOH and added to medium to a final concentration of 200 µM. Drugs were added to yeast suspension.
Immunohistochemistry
Samples were prefixed with 4% formaldehyde (solved in PBS) and washed three times (10 min) in PBS. Nuclei were stained with 50 μg Hoechst in glycerol/PBS (4:1) cover solution. During measurement, the same exposition time and magnification were used for all samples. After heatshock, testes were pre-dissected in PBS, then immunostaining was conducted according to the paper of Dahmann (2008). Anti-Phospho-Thor/4E-BP- (1:200, rabbit, Cell Signaling Technology, 2855,) was applied. The following secondary antibody was used: anti-Rabbit Alexa Fluor 488 (1:500, Life Technologies, A 11008). Antibodies were diluted in blocking solution. Nuclei were stained by Hoechst (0.1 mg/ml, Molecular Probes, 33342).
Fluorescent microscopy
Fluorescent images were captured with a Zeiss Axioimager Z2 upright microscope (with objectives Plan-NeoFluar 10 × 0.3 NA, Plan-NeoFluar 40 ×0.75 NA and Plan-Apochromat 63 × 1.4 NA) equipped with ApoTome, and a Nikon C2 confocal microscope (with objective 60 × Oil Plan APO VC NA = 1.45). AxioVision 4.82 and Jmage J 1.52c software were used to examine and evaluate data.
Electron microscopy
Adult flies were dissected in ice-cold PBS, and testes were fixed in a solution containing 2% formaldehyde, 0.5% glutaraldehyde, 3 mM CaCl2, and 1% sucrose in 0.1 M Na-cacodylate (pH 7.4; 30 min at room temperature); postfixed in 0.5% osmium tetroxide (45 min at room temperature) and in half-saturated aqueous uranyl acetate (30 min at room temperature), then dehydrated in graded series of ethanol, embedded in Durcupan (Sigma, 44610-1EA). White according to the manufacturer's instructions, and cured for 36 h at 60 °C. Ultrathin sections were stained with lead citrate for 5 min. Grids were analyzed in a JEOL JEM 1011 transmission electron microscope operating at 60 kV. Images were taken using an Olympus Morada 11 megapixel camera and iTEM software (Olympus). All reagents and materials used for electron microscopy were obtained from Sigma-Aldrich (Manzéger et al. 2021).
Western blot analysis
Western blot samples were prepared from 30 testes in 30 μl of Fly Lysis buffer and 30 μl 2×Laemmli buffer. 15 μl samples were run on 4–20% Mini-PROTEAN® TGX™ Gel and blotted onto Nitrocellulose Membrane (Kisker Biotech, 40520100). After blocking with 3% Milk Powder (BioRad 170-6404 /Blotting-Grade Blocker/), samples were dissolved in TBST, membranes were probed with anti-Tubulin (1:1000, mouse, Sigma T6199), anti-Ref(2)P/p62 (1:2000, rabbit gift of Gábor Juhász (Nagy et al. 2014), anti-Atg8a (1:2500, rabbit gift of Gábor Juhász), anti-P-S6k/Phosoho-RPS6KB1 (1:500, mouse, Cell Signaling Technology, 9206), anti-Atg13 (1:5000, rat gift of Gábor Juhász (Nagy et al. 2014), anti-mouse IgG alkaline phosphatase (1:1000, Sigma, A8438), anti-rat IgG alkaline phosphatase (1:1000, Sigma, A6066) and anti-rabbit IgG alkaline phosphatase (1:1000, Sigma, A3687) antibodies and developed by NBT-BCIP solution (Sigma, 72091).
Binding site analysis of Mad-Med heterodimer transcriptional factor
Sequences of Atg5 genomic regions from the Drosophila genus were obtained from FLYBASE (www.flybase.com). Conserved Mad-Med binding sites (Gao et al. 2005) were identified by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Potential binding sites were aligned with ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/).
PCR experiments
Apical part of 30 pairs testes were dissected from adult males in PBS, collected in TRI Reagent® solution (Zymo Research, R2050-1–50), and homogenized. RNA isolation was done according to the Direct-zol™ RNA MiniPrep kit (Zymo Research, R2050) protocol, which also includes a DNAse treatment. Reverse transcription was performed using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, K1621). The following primers were used in a semi-quantitative PCR analysis to amplify internal control: RpL32, forward 5ʹ-GCT AAG CTG TCG CAC AAA TGG-3ʹ and reverse 5ʹ-GTA GCC AAT GCC TAG CTT GTT C-3ʹ. For detection of bam forward 5ʹ-GGC GAT TGC TTA GAG CAC A-3ʹ and reverse 5ʹ-GGA AGC GCT CTA CTT TGG TG-3ʹ, Atg8a, forward 5ʹ- CAA TAC AAG GAG CAC GC-3ʹ and reverse 5ʹ-CGT GAT GTT CCT GGT ACA GGG A-3ʹ, Atg5 forward 5ʹ- GAC ATC CAA CCG CTC TGC-3ʹ reverse 5ʹ-TCG GCC AAA CTC TTC TTT TG-3ʹ.
Evaluation, statistics
The amount of autophagic structures in the fluorescence images was measured in the apical region. I defined a range (called apical region) within the peak of the testis and 55 µm from it. For statistical analysis of fluorescence microscopy, results were determined by using R Studio (Version 3.4.3). The distribution of samples (to decide normal or not) was tested with Lilliefors test. If it was normal, F-test was performed to compare variances. In cases when variances were equal, for clonal results, two-samples t test was used, otherwise one-sample t test was applied. For unequal variances was applied. In case of non-normal distribution, Mann-Whitney U test was performed. The level of significance was indicated as follows: *p < 0.05; **p < 0.01; ***p < 0.001. Gel images were analyzed by using ImageJ software. The intensity of saturation of pixels was corrected to the housekeeping control. Analysis of images taken of mCherry-Atg8a, Cathepsin-L and LysoTR-positive structures and amount of P-Thor was approached by using ImageJ program.
Results
Lack of lysosomal breakdown causes impairment in differentiation of SG cells and male fertility
Previous studies have shown that autophagy has an important role in cellular differentiation (Zhang et al. 2019). This information promoted us to measure lysosomal degradation in undifferentiated GSCs and their descendant, differentiated SG. Autophagic structures were labeled by mCherry-Atg8a reporter, which presents all vesicles from the forming phagophore to autolysosome (Lőrincz et al. 2017) and we stained the hub cells with anti-FacIII (which antibody labels only hub cells). The results indicate that more autophagic structures can be found further from FacIII-positive hub cells, and hub cells contain more acidic structures than the neighbor cells (Fig. 2 A). Visualized by electron microscopy, the appearance of hub cells is more dense (that might indicate the cytoplasm of the cell are more acidic) than the surrounding cells (Fig. S1 A). Staining with LysoTracker (LysoTR), which is a vital dye that marks lysosomes, late endosomes and autolysosomes (Manzéger et al. 2021), was applied to detect acidic compartments. Elevated activity of Bam was evident in SG, which are located farther from the hub (Chen et al. 2014). For this reason, the quantity of acidic and autophagic (mCherry-Atg8a) structures in cells expressing bam (through BamGal4-driven UAS-myr-GFP) is compared to the apical tip where BMP inhibits bam expression (Fig. 2 B-C’). Cells showing bam expression contained substantially more LysoTR- and mCherry-Atg8a-positive structures (Fig. 2 B’- C’). It is possible that the spatial distribution of autophagic structures is not even during the early spermatogenesis. Together, autophagy is more active in differentiated cells than in undifferentiated GSCs.
Autophagy is required for spermatogonial cells differentiation. A The distribution of autophagy is not even in the testis: Its level is decreased in GSCs as compared to the hub cells and SG. Autophagic compartments, such as phagophore, autophagosome and autolysosomes, are marked with mCherry-Atg8a (red) staining and hub cells were stained with anti-FasIII (green). In testis, GSCs were marked with dashed lines, located around the hub, the level of autophagy is decreased as compared to the neighboring cells. Testis is also marked by dashed lines. B-C’ Autophagic activity increases during spermatogonial differentiation. Hub cells are rich in acidic compartments, however, LysoTR-positive staining can be observed in the surrounding cells at a significantly lower level. The amount of autophagic and acidic structures increases at the site of Bam expression in contrast with the stem cell zone located around the hub. The expression area of bamGal4 can be seen in green (UAS-myristilated-Egfp-UAS-myr-GFP), star refers to the hub. B-B’ LysoTR refers to acidic compartments (in red). C–C’ Autophagic vesicles are marked with 3xmCherry-Atg8a (red). Bars represent ± S.D., *: P < 0.05; **: P < 0.01; ***: P < 0.001. For statistics, see the Materials and Methods. D-D’’ Atg genes are required for male fertility. Atg genes were silenced specifically in the germline, then animals were crossed with virgin females, and the fertility of males was tested. D To perform gene silencing, three different germline-specific Gal4 sources were used: nanosGal4 in the GSC and GB cells, bamGal4 in SG, and babGal4 in the complete testis. To attain the expression pattern of Gal4 systems, UAS-myrGFP was applied. All images were taken with the same exposure time. D’ The table shows the fertility of males during Atg silencing in percentage. D’’ Autophagy mutants show decreased fertility in percentage. E-E’ Aberrant testis morphology in autophagy-deficient background. In case of silencing Atg genes, animals become highly sterile. Thus, Atg downregulation was repeated and the morphology of testes was examined. In addition, Aut1 mutants were examined. E Autophagy deficient strains developed testes with aberrant morphology and decreased size. This phenotype, also known as bag of marbles, is observed in case of silencing bam. E’ The penetrance of morphological abnormalities is shown in percentage. F Anti-HTS was used to label spectrosomes (green). We observed much more spectrosomes in Aut1 mutant to the control. Cell nuclei were stained with Hoechst.
In further experiments, silencing of Atg genes was driven by three different Gal4 constructs, which had different expression patterns in the male germline, and male fertility was investigated. nanos-Gal4 is active only in GSCs and two-cell-gonialblasts (Gb), bam-Gal4 is expressed in both two-cell-Gb and 16-cell-SG, while bab-Gal4 is expressed in the entire germline (Fig. 1 D) (White-Cooper et al. 2012). Figure 2 D shows the expression pattern and intensity of these Gal4 strains. bab-Gal4 is expresssed in the entire germline, although its expression intensity is decreased as compared to the other two Gal4 sources. According to our results, silencing autophagy-related (Atg) and Vha16 (V-type proton ATP-ase 16 kDa proteolipid subunit) lysosomal proton pump genes by RNA-interference (TRiP) significantly decreased male fertility (Fig. 2 D’). Sterility was most evident in animals silenced by the Bam-Gal4 driver (Fig. 2 D’) in the case of Atg1-TRiP, Atg2-TRiP, Atg5-TRiP, Vha16-TRiP, and Atg14-TRiP. Sterility in Atg7∆77/Atg7∆14 and Aut110(Atg3) loss-of-function mutants was also detectable (Fig. 2 D’’). Given the fact that silencing of Atg5 and Vha16 through bam-Gal4, as well as Aut110, mutants showed the highly penetrant sterile phenotype, we assessed the impact of gene silencing on the testis morphology and compared it with the phenotype caused by Bam deficiency (bam-TRiP) (Fig. 2 E-E’). Downregulation of bam was previously shown to lead to a bag-of-marbles (BOM) phenotype, which refers to SG-clusters containing a large number of cells (Chen et al. 2014). Cells within SG cannot enter into meiosis, rather they stucke at mitosis, resulting in large cysts. Beside Atg5, downregulation of Vha16 and a mutation in Aut1 also caused an abnormal morphology of the testis, in which BOM-like cysts could been distinguished (Fig. 2 E-E’). We labeled control and autophagy-deficient testes with anti-HTS labeling spectrosomes and we observed much more spectrosomes in Aut1 mutant and Vha16 silenced samples compared to the control. These results also indicate that a partial inhibition of meiosis can occur in autophagy-deficient germline (Fig. 2 F and Fig. S2 A).
Bafilomycin and wortmannin inhibit autophagy at different stages of the process. Bafilomycin suppresses vesicle fusion and lysosome acidification (Mauvezin et al. 2015), whereas wortmannin interferes with the Vps34 vesicle nucleation complex (Moskalev et al. 2010). We found that treating animals with bafilomycin and wortmannin similarly causes a BOM-like phenotype in the affected testis (Fig. S2 B-B’). Based on these results, we conclude that autophagy is active in differentiating SG, and that defects in the process cause a BOM-like phenotype in testis morphology. The latter feature is a characteristics of mutant animals deficient in Bam function.
BMP signaling inhibits autophagy in GSCs
In germline cells, in which the BMP signaling pathway is active, autophagy was detected to operate at basal levels (Fig. 2 A-C’). This may raise the possibility that BMP signaling can inhibit the autophagic processes in these cells. To address this issue, we first performed an in silico analysis of Atg genes and their upstream regulatory sequences to search for conserved binding sites of the Mad-Med transcription factors, effectors of BMP signaling. We found a potential Mad-Med binding site within the fifth intronic sequences of the Atg5 coding region (Fig. S3 A). Next, consistent with this finding, Atg5 expression was increased in testes downregulated for mad (mad-TRiP) and lowered in tkvQ253D.Nb gain-of-function mutants (Fig. S3 B-C’). Based on these data, one can suppose that the expression of Atg5 is negatively regulated by the BMP pathway. Further studies should reveal a direct interaction between Mad-Med and the Atg5-specific binding element. Downregulation of mad, which represses BMP signaling, markedly increased transcript levels of both bam and Atg8a (Fig. S3 B). Indeed, in GSCs, bam activity was previously demonstrated to be under the control of BMP signaling (Kawase et al. 2004). However, we could identify no conserved Mad-Med binding site in the Atg8a locus. We suggest that repression of certain Atg genes in GSCs is achieved by a mechanism that is distinct from the transcriptional regulation of BMP signaling.
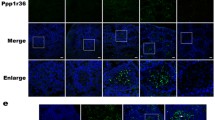
We also applied autophagy-related fluorescent markers, LysTR and mCherry-Atg8a (mCh-Atg8a), to test the regulatory effect of BMP signaling on autophagy (Fig. 3 A-B’). In this set of experiments, we silenced the mad gene (mad-TRiP), overexpressed dpp ligand, and used a gain-of-function tkvQ253D.Nb and loss-of-function tkv4alleles of tkv receptor. For these genetic interventions (in the case of USA- constructs), nanos-Gal4 was used which is expressed in both GSCs and Gbs. We found that the number of LysoTR- and mCh-Atg8a-positive structures increases in testes treated with mad-TRiP (Fig. 3 A-B’). In contrast, hyperactivation of BMP signaling by overexpressing tkvQ253D.Nb and dpp significantly decreased the number of autophagic structures (Fig. 3 A’ and B’). It is worth noting that ectopic expression of Dpp led to a BOM phenotype (Fig. S3 D). Clones cells were created by tkv4 loss-of-function mutants (Fig. 3 C–C’), in which the amount of acidic compartments was determined. Homozygous tkv4 mutant cells (area at the tip of the testis, not in green-the clonal region contains both somatic and germline cells) displayed increased levels of LysoTR-positive structures, as compared with control (testes with no mutant clone cell were used as control). In this case, we detected no increase in the number of lysosomes and autolysosomes in the apical area of the testis. Similarly to previous cases (Fig. 2 A-B), a decreased number of LysoTR-positive structures was detected at the apical site of these testes (Fig. 3 C). Connection between autophagy and BMP signaling was further analyzed by measuring p62 levels, which serves as an adaptor protein for autophagy. p62 is also degraded by autophagic breakdown, so its levels inversely correlate with autophagic activity. The fly ortholog of mammalian p62 is Ref(2)P (Nezis et al. 2008). The amount of Ref(2)P/p62 decreased in testes treated with mad-TRiP, while increased in the tkv Q253D.Nb gain-of-function genetic background (Fig. 3 D-D’). Taken together, BMP signaling appears to inhibit autophagy in GSCs, and this inhibition may be originated in part from the negative transcriptional regulation of Atg5.
The BMP pathway inhibits autophagy. A-B’ Ectopic activation of BMP signaling decreases the amount of autophagic and acidic compartments. The effect of BMP signaling on autophagy was investigated through the alteration of expression of the transcription factor mad, Type I receptor tkv and the ligand dpp. A-B For microscopic analysis of autophagy, A and LysoTracker Red (LysoTR – acidic components) B) and mCherry-Atg8a (phagophore, autophagosome and autolysosome) fluorescent markers were applied. The amount of acidic and autophagic compartments increased as compared to the control, as a result of mad downregulation (mad-TRiP). Gain-of-function allele of tkv resulted in decreased levels of autophagy. UAS-transgenes were driven by nanosGal4. The same phenomenon was obtained by overexpressing dpp ligand. These testes show significantly decreased amounts of LysoTR- and mCherry-Atg8a-positive compartments. In Dpp overexpressing animals, LysoTR-positive components were not detected. C–C’ The amount of acidic compartments are increased in tkv loss-of-function mutants. Amounts of LysoTR (red) increase in clonal cells (not green cells drawn around with dashed lines) of loss-of-function tkv mutant strains, as compared to control cells (green). Distribution of acidic structures was assessed in testes where control cells were not induced as control. Tkv mutant clonal cells show intense LysoTR staining at the apical site of the testis. D-D’ Ref(2)P/p62 is a substrate for autophagy. The quantity of the protein was determined by Western blot technique. D’ The level of p62 decreases in samples with reduced mad activity and increases in animals overexpressing the gain-of-function allele of tkv. αTub84B was used as an internal control. Bars represent ± S.D., *: P < 0.05; **: P < 0.01; ***: P < 0.001. For statistics, see the Materials and Methods
The bam differentiation factor enhances autophagy
The number of autophagic structures became elevated in the area where Bam is expressed as compared to the posterior stem cell niche zone (Fig. 2 B-C’). Furthermore, inhibition of Atg genes led to a BOM phenotype (Fig. 2 E-E’). BMP signaling blocks autophagy (Fig. 3) and is known to repress bam (Bangi et al. 2006). This prompted us to ask whether Bam influences autophagic activity. To this end, we overexpressed bam by a heat-shock promoter (hs-bam) and examined the effect of this intervention on autophagy (Fig. 4 A-C and Fig. S4 A-C). In case of bam overexpression, the number of LysoTR- and mChAtg8a-positive structures increased (Fig. 4 A-B’). We also examined whether these changes occur in the absence of hs-bam transgene. None of the markers showed alteration in this genetic background (thus our results are independent of heat shock) (Fig. S4 A-B’). So, an increase in the amount of autophagic structures was due to bam overexpression. We confirmed these results by performing a Western blot analysis, in which p62 and Atg8a protein levels were determined in control vs. bam-overexpressing genetic backgrounds (Fig. 4 C and Fig. S4 C). p62 levels lowered when Bam was hyperactivated. This indicates an enhanced autophagic activation in testes with excessive Bam function. The level of Atg8a-I and -II isoforms increased during bam overexpression (Fig. 4 C). The ratio of Atg8a-I and -II was also different relative to each other contrary to control (where the level of Atg8a-II was significantly higher than Atg8a-I). When Bam was hyperactivated, difference between the two Atg8a isoforms was decreased. Atg8a-II is anchored to the autophagic membrane and serves as a substrate for autophagy, while Atg8a-I is a soluble protein in the cytoplasm (Mizushima et al. 2007). Decreased ratio of the two isoforms also refers to elevated autophagy. Downregulation of bam decreased the amount of lysosomal vesicles, but increased the number of autophagic structures (Fig. 4 D-E’). Elevation of Atg8a-II and p62 protein levels was demonstrated by a Western blot analysis (Fig. 4 F and Fig. S4 D). These results suggest that autophagy is inhibited after the stage of Atg8a lipidation, which can be caused by defects in vesicle fusion or acidic degradation. The effectivity of hs-bam and bam-TRiP constructs was determined by semi-quantitative PCR (Fig. S4 E). These data showed that bam transcript levels elevate in testes with Bam hyperactivity, but lower in testes downregulated for bam. Then, we assessed changes in Atg5 mRNA levels in these genetic backgrounds with semiquantitative-PCR and found that Bam downregulates Atg5 (Fig. S4 E). While homozygous bam∆86 mutants are unviable, heterozygous bam∆86 /Df(3R)FFD−0089346 mutants are viable (that suggests the bam∆86 mutant flies contain a background mutation in a differentdifferent chromosomal position, which mutation causes the lethal phenotype) and exhibit morphological abnormalities (Fig. S5 A). Similar to bam-TRiP animals, heterozygous bam∆86 /Df(3R)FFD−0089346 mutants contained fewer amounts of LysoTR-positive acidic compartments as compared to control (Fig. S5 B-B’). In electron microscope images, amphisomal structures are however accumulated in bam∆86 /Df(3R)FFD−0089346 mutants (Fig. S5 C-C’). Cathepsin-L is a lysosomal hydrolase, and its levels positively correlate with the amount of lysosomes and autolysosomes (McKearin and Ohlisten 1995). We found that the number of Cathepsin-L-positive vesicles greatly decreases in testes expressing bam-TRiP (Fig. 4 G-G’). In sum, Bam is capable of stimulating autophagy in the Drosophila testis.
The differentiation-promoting factor Bam has an autophagy-activating effect in the male germ line. A-B’. Due to the overexpression of bam, the measure of acidic (A-A’) and the mCherry-Atg8a-positive (B-B’) structures was increased as compared to the control. C The amount of Ref(2)P/p62 and Atg8-II proteins was analyzed by Western blot. Downregulation of bam (for one day) decreases p62 levels and increases Atg8a-II levels. Thus, Bam activates autophagy (A-C). D-E’) Bam downregulation lowers the amount of LysoTR-positive compartments (bam-TRiP is driven by bamGal4) and increases the number of mCherry-Atg8a-positive particles (E-E’). F) Upon bam silencing, the amount of p62 and Atg8a-II proteins was increased. G-G’) In the germ line, the number of cathepsin-L-positive structures (lysosomes and autolysosomes) is lowered in response to bam deficiency. These results suggest that Bam influences autophagy after Atg8a lipidation (at vesicle fusion or autolysosomal degradation). Nuclei were stained by Hoechst (blue)Bars represent ± S.D., *: P < 0.05; **: P < 0.01; ***: P < 0.001. For statistics, see the Materials and Methods
Bam regulates lysosomal breakdown through the Tut-Bam-Bgcn complex
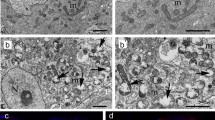
The differentiation-promoting factor Bam functions in the Tut-Bam-Bgcn complex (Chen et al. 2014). Therefore, we next downregulated tut and bgcn and detected a BOM phenotype in the treated animals (Fig. S6 A). In these animals, the amount of LysoTR- and Cathepsin-L-positive structures was also decreased (Fig. S6 B-C’), whereas Ref(2)P/p62 levels became significantly enhanced (Fig. 5 A-A’). In case of tut-TRiP, the amount of Atg8a-II-positive foci was heavily increased, too (Fig. 5 A). These results indicate that the Tut-Bam-Bgcn complex exerts its effect on autophagy after Atg8a lipidation. Electron microscopy was further applied to examine germline cells defective for Tut or Bam function. The analysis revealed numerous late endosomes (multivesicular bodies) and amphisomes, but not lysosomal compartments in the samples (Fig. 5 B and Fig. S6 D). Tut-Bam-Bgcn deficiency is hence associated with impaired both autophagic and endocytic degradation. To decide whether vesicle fusion or lysosomal breakdown is compromised in these animals, we compared the phenotype of bam and tut deficiency with Vha16-RNAi and syx17LL mutants. Vha16 deficiency leads to an abnormal pH within the lysosomal lumen (i.e., lysosomal hydrolases cannot function), whereas inactivating mutations in syx17 blocks the autophagosome-lysosome fusion (Takáts et al. 2014). Accumulation of lysosomes and autophagosomes was noticed lacking Syx17 (S.Fig. 6 D’). However, Vha16-TRiP resulted a phenotype similar to the silencing of tut and bam (Fig. 5 B). Thus, we could not detect lysosomes (next to the aphisomes and multivesicular bodies) in the absence of the Tut-Bam-Bgcn complex. Given the fact that, in addition to autophagy, the endocytotic degradation pathway is also affected, distribution of Rab5-positive structures was also examined in tut-TRiP testes (Fig. 5 C–C’’). Rab5 indicates early endosomal vesicles (Wucherpfennig et al. 2003), the size of which is enlarged in testes lacking Tut function. This suggests that endosomal maturation is impaired in the Tut-Bam-Bgcn defectice testis. Therefore, the Tut-Bam-Bgcn complex affects both autophagy and endosomal degradation (heterophagy) through regulating lysosomal breakdown.
Bam regulates lysosomal degradation in the germ line through its role in the Tut-Bam-Bgcn complex. A-A’ Genes encoding two members of the Bam complex, Tut and Bgcn, were silenced post-transcriptionally. Downregulation increases p62 levels. B In the testis, bam and tut (downregulation of them caused a more severe phenotype than that of bgcn) functions were examined ultrastructurally by electron microscopy. Downregulation of these genes increases the number of amphisomes and multivesicular bodies in the spermatogonia, but not of acidic dense components (lysosomes and autolysosomes). Treatment was performed in Vha16-TRiP (vacuolar proton-ATPase) samples, in which large amphisomes and late endosomes (multivesicular bodies) accumulate. C Downregulation of tut increases the size, but not the number, of Rab5-positive endosomal structures. Thus, the Tut-Bam-Bgcn complex plays a role in the acidification of lysosomes. Bars represent ± S.D., **: P < 0.01. For statistics, see the Materials and Methods
The Tut-Bam-Bgcn complex regulates lysosomal degradation through activating TORC1. A Atg13, one of the member of the Atg1 initiation complex, is hiperphosphorilated (p-Atg13) upon TORC1 inhibition. p-Atg13 levels increase in testes deficient in bam and tut. Phosphorylation of the ribosomal protein S6k (p-S6k) is mediated by TORC1. BamGal4 driver was used for silencing. αTub84B was applied as an internal control. B The expression of Bam (by BamGal4-driven UAS-myr-GFP) was examined by anti-P-Thor/4EBP antibody staining (phosphorylation of Thor/4EBP is under the control of TORC1). The level of anti-P-Thor/4EBP shows a peak in the spermatocytes, and its spatial distribution is similar to Bam expression. C–C’ The amount of p-Thor/4EBP was significantly decreased compared to the control. Bars represent ± S.D., **: P < 0.01. For statistics, see the Materials and Methods. D Morphology of the apical part of the testes is abnormal in animals downregulated for Tor which refers to damages in early spermatogenesis. Nuclei were stained by Hoechst vital dye. E-E’ The amount of Ref(2)P/p62 decreases in samples overexpressing Bam for one day while increases in samples overexpressing Bam for three days. F Blocking the BMP signaling pathway makes possible the expression of Bam and activates both TORC1 complex and autophagy
The Tut-Bam-Bgcn complex modulates with TORC1 to control SG differentiation
The Atg1 autophagy induction complex can be activated by inhibiting TORC1 (Target Of Rapamycin kinase Complex 1) (Alers et al. 2012). The inhibition of TORC1 promotes the hyperphosphorylation of Atg13 (p-Atg13), an integrant part of the complex (Chang and Neufeld 2009). TORC1 is localized to lysosomes when functioning (Li et al. 2019). In the case of prolonged activation of autophagy, TORC1 may also have a role in lysosome biogenesis. We were curious whether the Tut-Bam-Bgcn complex affects TORC1 activity. The phosphorylated state of Atg13, which occurs through S6k (phosphorylated ribosomal protein S6 kinase-P-S6k) and Thor/4E-BP1 (thor phosphorylated 4E (eIF4E)-binding protein 1/ P-Thor), indicates TORC1 activity (Wullschleger et al., 2006; Fischer et al., 2015). Level of P-Atg13 and P-S6k was examined by a subsequent Western blot analysis (Fig. 6 A and Fig. S7 A-A’). We found that P-Atg13 levels elevate, while P-S6k amounts decrease (under a detectable treshold) in bam- and tut-RNAi backgrounds (Fig. 6 A and Fig. S7 A’). However, we observed a decreased amount of P-Atg13 upon bam overexpression (Fig. S7 A-A’). Immunhistochemistry was conducted to determine the distribution of phosphorylated Thor/4E-BP in the testis, and results showed a pattern similar to bam expression at the apical site of the testes (Fig. 6 B). Downregulation of bam and tut resulted in decresed levels of P-Thor in this organ (Fig. 6 C-C’ and Fig. S7 B-B’). Based on these results, one can propose that the Tut-Bam-Bgcn complex is neccessary for the appropriate activity of TORC1 during spermatogenesis. We also examined whether TORC1 activity has a role in this differentiation paradigm. Tor downregulation led to testes with a BOM-like phenotype (penetrance 20/40) (Fig. 6 D). We observed a similar phenotype (penetrance 9/28) in case of applying Rapamycin, a pharmacological inhibitor of TORC1 (Fig. S7 C). Finally, we compared autophagic activity between samples exposed to one-day-long and three-day-long bam expression (Fig. 6 E-E’ and Fig. S7 D). While the one-day-long regimen enhanced autophagy, the three-day-long treatment greatly increased the amount of Ref(2)P/p62, i.e., it suppressed autophagy. Together, the Tut-Bam-Bgcn complex is able to activate TORC1, which is in turn required for spermatogonial differentiation and promotes the meiotic program.
Discussion
Results presented in this study suggest that autophagy is required to early spermatogenesis in Drosophila (Fig. 1). Inhibition of autophagy caused male infertility and differentiational defects similar to bam mutation: Cells were not able to enter meiosis, but continued mitosis, thus abnormal sized spermatogonia cysts were produced. In GSCs, as compared to SG that were differentiated from them, we managed to detect a more intense autophagy activity in the later cells. For this reason, it is plausible that autophagy is required for differentiation of SG (Fig. 1). (Zhao et al. 2018) examined the distribution of autophagy in the germline of Drosophila females and showed that autophagy was decreased in GSCs as compared to their descendants (cytoblasts and cystocytes) (Zaho et al. 2018), similarly as it occurs in males. The connection between autophagy and differentiation is intensively studied previously. However, the accurate functions of autophagy in this differentiation paradigm are still unknown. During metamorphosis of Drosophila, superflous larval tissues of pupae, such as fat body or salivary gland, are breakdown to transfer it into a mature imago. In this tissue degradation process, autophagy plays a critical role. Activation of Type I PI3 K (through activation of Ras, Akt or Dp110 – a type I PI3 K contact reaction subunit) inhibits the degradation of salivary gland in pupae.
During autophagy-dependent degradation of salivary gland, both Type I PI3 K and TORC1 are inhibited (Tracy and Baehrecke 2013). In Drosophila midgut, programmed cell death and a balance between the type I PI3 K pathway and autophagy are crucial for normal development. The pathway is known to inhibit autophagy to adjust the timing of cell death (Denton et al. 2019). Mutations in Atg genes result in increased expression of PI3K and inhibition of developmental degradation in the midgut (Tracy and Baehrecke 2013). In nematodes (C. elegans), the maternal inheritance of mitochondria is a passive process, since the paternal originated mitochondria are degraded by selective autophagy (Yang and Zhang 2014). In the fruitfly, contrary to larval tissues, imaginal primordia (they develop to form the organs of the imago) are not degraded during metamorphosis, however, differentiation of these tissues requires autophagy. During the development of the compound eye of Drosophila, an intense autophagic activity can be observed in the eye disks. Inhibition of autophagy results in impaired eye development (Billes et al. 2018). In this case, increased autophagy does not lead to cell death. Therefore, the role of developmental autophagy can vary depending on the origin of tissue.
Autophagy has a crucial role in maintaining stem cells and in their differentiation (Zhang et al. 2019). A decreased level of autophagy was detected in GSCs relative to SGs. The Gbb/BMP (BMP) pathway mediates GSC maintaining (in part by inhibiting bam expression to maintain the stemness state) (Kawase et al. 2004). Here, we investigated the potential role of the BMP pathway in autophagy regulation in the apical site of the germ line. According to our results, BMP signaling inhibits autophagy in this paradigm (Fig. 3). In the degradation of Drosophila gut cells, autophagy plays a critical role; the lack of Dpp, which serves as a ligand for the BMP pathway, results in autophagy hiperactivity and early cell death (Denton et al. 2019). Thus, the BMP signaling pathway also inhibits autophagy in larval midgut cells.
According to our understanding, the Tut-Bam-Bgcn complex activates autophagy during differentiation of SG (Fig. 5). Overexpression of Bam caused increased levels of autophagy, whereas inhibition of the Tut-Bam-Bgcn complex resulted in a significantly compromised degradation of autophagic cargos (Figs. 4–5). It was previously shown in the Drosophila female germ line that lack of. In the present study, we demonstrated that inhibition of the Bam-Tut-Bgcn complex results in impaired lysosomal degradation. Abundant autophagic and endosomal structures could be seen by fluorescent microscopy and ultrastructural assays, nevertheless lysosomes were missing from the cells (Figs. 4–5 and S.Figs. 4–6). Based on electron microscopic images of the female germ line, lysosomal degradation can be affected in the early oogenesis. In Bam deficient mutants, autophagosomes accumulate, however, autolysosomes and lysosomes are failed to be detected (Zaho et al. 2018). However, Gm114 is described as Bam orthologue in mice with a similar expression pattern, its function is not yet known (Tang et al 2008).
We also examined the potential connection between the Tut-Bam-Bgcn differentiation complex and lysosomal degradation. To our surprise, inhibition of the differentiation complex intererred with TORC1 activity, which resulted in testes with an abnormal morphology being similarly to that found bam mutants (Fig. 6). It is known that TORC1 blocks autophagy during differentiation of stem cells (Chen et al. 2008). In mammals, hematopoietic stem cells were shown to maintain their undifferentiated state only when autophagy is present. It was previously observed during differentiation of hematopoietic stem cells that the activity of mTOR increases (Chen et al. 2008; Owusu-Ansah and Banerjee 2009). Inhibition of mTORC1 enhances autophagy, which in turn blocks the differentiation of hematopoietic cells (Hoshii et al. 2012). However, TORC1 can induce long-term (continuous starvation for 18–24 h) autophagy during starvation. Li et al. 2019 investigated the role of TORC1 during long-term starvation in various model systems, such as larval fat bodies of Drosophila, mouse kidney, fish, bird and mammalian cells. During prolonged starvation, sustainment of autophagy is mediated by periodical re-activation of TORC1. This activation of TORC1 includes stimulated vesicle and tubule formation from autophagosomal membranes. These lysosomal lipid components later on generated new functional lysosomes, which were indispensable for high levels of autophagy (Li et al. 2019). Results presented here indicated that prolonged overexpression of Bam results in autophagy inhibition (Fig. 6). We hypothetize that periodic activity of TORC1 may have a similar effect on germ cell differentiation than that of long-term starvation. TORC1 can enhance differentiation without modifying autophagy, because it also influences processes such as cell growth, proliferation, mitochondria production and cell migration (Serbus et al. 2015).
We suggest that autophagy is required for the differentiation of germ line SG. Autophagy is under the regulation of BMP signaling in germ line stem cells, while in SG (where BMP is inactive), the Tut-Bam-Bgcn complex induces autophagy. The Tut-Bam-Bgcn complex is necessary for increased level of long-term autophagy during differentiation. Presumably, that complex supplies new primordial lysosome vesicles (recycling of lysosomal membranes) for digestion. This long-term elevated level of autophagy is partly mediated by TORC1 (Fig. 6 F). Our results may help to understand regulatory differences between developmental (long-term) and stress-induced autophagy.
Conclusion
Here, we investigated the role of autophagy in the differentiation of the male germ line. Our results show that autophagy is indispensable for the appropriate differentiation of diploid SG. We also found that in stem cells, autophagic breakdown is under the inhibition of the BAM signaling pathway, while in the stem cell daughters, the process is induced by BAM signaling. Bam is able to regulate acidic degradation and autophagy through activating TORC1.Thus, we explored a novel role for TORC1 in cellular differentiation.
Data Availability
Our journal mandates that all manuscripts include a Data Accessibility section which states where the article’s supporting data and digital research materials can be accessed. The data should be included in the reference list of the paper, too. We encourage authors to deposit their data in external, publicly accessible repositories, where possible. If the data have been deposited in an external repository, this section should list the database, accession number and link to the DOI for all data from the article that have been made publicly available. If the data are included in the article’s Supplementary Material, this should be stated here, for instance: The datasets supporting this article have been uploaded as part of the Supplementary Material.
References
Alers S, Löffler AS, Wesselborg S, Stork B (2012) The incredible ULKs. Cell Commun Signaling. https://doi.org/10.1186/1478-811X-10-7
Araujo H, Fontenele MR, da Fonseca RN (2011) Position matters: Variability in the spatial pattern of BMP modulators generates functional diversity. Genesis 49:9. https://doi.org/10.1002/dvg.20778
Bangi E, Wharton K (2006) Dpp and Gbb exhibit different effective ranges in the establishment of the BMP activity gradient critical for Drosophila wing patterning. Dev Biol 295:1. https://doi.org/10.1016/j.ydbio.2006.03.021
Billes V, Kovács T, Manzéger A, Lőrincz P, Szincsák S, Regős Á, Kulcsár PI, Korcsmáros T, Lukácsovich T, Hoffmann G, Erdélyi M, Mihály J, Takács-Vellai K, Sass M, Vellai T (2018) Developmentally regulated autophagy is required for eye formation in Drosophila . Autophagy 14:9. https://doi.org/10.1080/15548627.2018.1454569
Blagosklonny MV (2019) Rapamycin for longevity: opinion article. Aging 11(19):8048. https://doi.org/10.18632/aging.102355
Blatt P, Wong-Deyrup SW, McCarthy A, Breznak S, Hurton MD, Upadhyay M, Bennink B, Camacho J, Lee MT, Rangan P (2021) RNA degradation is required for the germ-cell to maternal transition in Drosophila . Current Biology CB 31(14):2984-2994.e7. https://doi.org/10.1016/j.cub.2021.04.052
Bonfini A, Dobson AJ, Duneau D, Revah J, Liu X, Houtz P, Buchon N (2021) Multiscale analysis reveals that diet-dependent midgut plasticity emerges from alterations in both stem cell niche coupling and enterocyte size. ELife. https://doi.org/10.7554/eLife.e64125
Boya P, Codogno P, Rodriguez-Muela N (2018) Autophagy in stem cells: repair, remodelling and metabolic reprogramming. Development 145:4. https://doi.org/10.1242/dev.146506
Chang YY, Neufeld TP (2009) An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell 20:7. https://doi.org/10.1091/mbc.e08-12-1250
Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, Liu Y, Zheng P (2008) TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med 205:10. https://doi.org/10.1084/jem.20081297
Chen D, Wu C, Zhao S, Geng Q, Gao Y, Li X, Zhang Y, Wang Z (2014) Three RNA binding proteins form a complex to promote differentiation of germline stem cell lineage in Drosophila . PLoS Genet 10:11. https://doi.org/10.1371/journal.pgen.1004797
Cheng J, Tiyaboonchai A, Yamashita YM, Hunt AJ (2011) Asymmetric division of cyst stem cells in Drosophila testis is ensured by anaphase spindle repositioning. Development 138:5. https://doi.org/10.1242/dev.057901
Cunningham KM, Maulding K, Ruan K, Senturk M, Grima JC, Sung H, Zuo Z, Song Helen, Gao J, Dubey S, Rothstein JD, Zhang K, Bellen HJ, Lloyd TE (2020) TFEB/Mitf links impaired nuclear import to autophagolysosomal dysfunction in C9-ALS. eLife. https://doi.org/10.7554/eLife.59419
Dahmann C (2008) Drosophila - Methods and Protocols. Methods Mol Biol. https://doi.org/10.1128/mSphere.00698-21
Dansereau DA, Lasko P (2008) RanBPM regulates cell shape, arrangement, and capacity of the female germline stem cell niche in Drosophila melanogaster. J Cell Biol 182:5. https://doi.org/10.1083/jcb.200711046
de Cuevas M, Matunis EL (2011) The stem cell niche: Lessons from the Drosophila testis. Development 138:14. https://doi.org/10.1242/dev.056242
Denton D, Xu T, Dayan S, Nicolson S, Kumar S (2019) Dpp regulates autophagy-dependent midgut removal and signals to block ecdysone production. Cell Death Differ 26:4. https://doi.org/10.1038/s41418-018-0154-z
Fischer KE, Gelfond JAL, Soto VY, Han C, Someya S, Richardson A, Austad SN (2015) Health effects of long-term rapamycin treatment: The impact on mouse health of enteric rapamycin treatment from four months of age throughout life. PLoS ONE 10:5. https://doi.org/10.1371/journal.pone.0126644
Flores HA, Bubnell JE, Aquadro CF, Barbash DA (2015) The Drosophila bag of marbles Gene Interacts Genetically with Wolbachia and Shows Female-Specific Effects of Divergence. PLoS Genet 11:8. https://doi.org/10.1371/journal.pgen.1005453
Follansbee TL, Gjelsvik KJ, Brann CL, McParland AL, Longhurst CA, Galko MJ, Ganter GK (2017) Drosophila nociceptive sensitization requires BMP signaling via the canonical SMAD pathway. J Neurosci 37(35):8524–8533
Gao S, Steffen J, Laughon A (2005) Dpp-responsive silencers are bound by a trimeric Mad-Medea complex. J Biol Chem 280:43. https://doi.org/10.1074/jbc.M506882200
Gönczy P, Matunis E, DiNardo S (1997) bag-of-marbles and benign genial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Development 124:21. https://doi.org/10.1242/dev.124.21.4361
Hamaratoglu F, Affolter M, Pyrowolakis G (2014) Dpp/BMP signaling in flies: From molecules to biology. Semin Cell Dev Biol. https://doi.org/10.1016/j.semcdb.2014.04.036
Hayashi Y, Kobayashi S, Nakato H (2009) Drosophila glypicans regulate the germline stem cell niche. J Cell Biol 187:4. https://doi.org/10.1083/jcb.200904118
Hoshii T, Tadokoro Y, Naka K, Ooshio T, Muraguchi T, Sugiyama N, Soga T, Araki K, Yamamura KI, Hirao A (2012) mTORC1 is essential for leukemia propagation but not stem cell self-renewal. J Clin Investig 122:6. https://doi.org/10.1172/JCI62279
Insco ML, Leon A, Tam CH, McKearin DM, Fuller MT (2009) Accumulation of a differentiation regulator specifies transit amplifying division number in an adult stem cell lineage. Proc Natl Acad Sci USA 106:52. https://doi.org/10.1073/pnas.0912454106
Insco ML, Bailey AS, Kim J, Olivares GH, Wapinski OL (2012) A self-limiting switch based on translational control regulates the transition from proliferation to differentiation in an adult stem cell lineage. Cell Stem Cell 11:689–700. https://doi.org/10.1016/j.stem.2012.08.012
Juhász G, Csikós G, Sinka R, Erdélyi M, Sass M (2003) The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett 543(1–3):154–158. https://doi.org/10.1016/s0014-5793(03)00431-9
Kawase E, Wong MD, Ding BC, Xie T (2004) Gbb/Bmp signaling is essential for maintaining germline stem cells and for repressing bam transcription in the Drosophila testis. Development 131:6. https://doi.org/10.1242/dev.01025
Kondo S, Takahashi T, Yamagata N, Imanishi Y, Katow H, Hiramatsu S, Lynn K, Abe A, Kumaraswamy A, Tanimoto H (2020) Neurochemical Organization of the Drosophila brain visualized by endogenously tagged neurotransmitter receptors. Cell Rep 30(1):284-297.e5. https://doi.org/10.1016/j.celrep.2019.12.018
Kovács T, Billes V, Komlós M, Hotzi B, Manzéger A, Tarnóci A, Papp D, Szikszai F, Szinyákovics J, Rácz Á, Noszál B, Veszelka S, Walter FR, Deli MA, Hackler L, Alfoldi R, Huzian O, Puskas LG, Liliom H, Vellai T (2017) The small molecule AUTEN-99 (autophagy enhancer-99) prevents the progression of neurodegenerative symptoms. Sci Rep. https://doi.org/10.1038/srep42014
Kuhn H, Sopko R, Coughlin M, Perrimon N, Mitchison T (2015) The Atg1-Tor pathway regulates yolk catabolism in Drosophila embryos. Development (cambridge, England) 142(22):3869–3878. https://doi.org/10.1242/dev.125419
Li W, Zhang L (2019) Regulation of ATG and autophagy initiation. Adv Exp Med Biol. https://doi.org/10.1007/978-981-15-0602-4_2
Li Y, Zhang Q, Carreira-Rosario A, Maines JZ, McKearin DM, Buszczak M (2013) Mei-p26 cooperates with bam, bgcn and sxl to promote early germline development in the Drosophila ovary. PLoS ONE 8:3. https://doi.org/10.1242/dev.077412
Li M, Hu X, Zhang S, Ho MS, Wu G, Zhang L (2019) Traffic jam regulates the function of the ovarian germline stem cell progeny differentiation niche during pre-adult stage in Drosophila. Sci Rep 9(1):10124. https://doi.org/10.1038/s41598-019-45317-6
Lim C, Gandhi S, Biniossek ML, Feng L, Schilling O, Urban S, Chen X (2015) An aminopeptidase in the Drosophila testicular niche acts in germline stem cell maintenance and spermatogonial dedifferentiation. Cell Rep 13(2):315–325. https://doi.org/10.1016/j.celrep.2015.09.001
Manzéger A, Tagscherer K, Lőrincz P, Szaker H, Pilz P, Kméczik R, Csikós G, Erdélyi M, Sass M, Kovács T, Vellai T, Billes VA (2021) Condition-dependent functional shift of two Drosophila Mtmr lipid phosphatases in autophagy control. Autophagy 28:1–19. https://doi.org/10.1080/15548627.2021.1899681
Mauvezin C, Nagy P, Juhász G, Neufeld TP (2015) Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun. https://doi.org/10.1038/ncomms8007
McKearin D, Ohlstein B (1995) A role for the Drosophila Bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development 121:9. https://doi.org/10.1242/dev.121.9.2937
Mizushima N, Levine B (2010) Autophagy in mammalian development and differentiation. Nat Cell Biol 12:9. https://doi.org/10.1038/ncb0910-823
Mizushima N, Yoshimori T (2007) How to interpret LC3 immunoblotting. Autophagy 3:6. https://doi.org/10.4161/auto.4600
Moskalev AA, Shaposhnikov M (2010) Pharmacological inhibition of phosphoinositide 3 and TOR kinases improves survival of Drosophila melanogaster. Rejuvenation Res 13:2–3. https://doi.org/10.1089/rej.2009.0903
Nagy P, Kárpáti M, Varga Á, Pircs K, Venkei Z, Takáts S, Varga K, Érdi B, Hegedus K, Juhász G (2014) Atg17/FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in Drosophila. Autophagy 10:3. https://doi.org/10.4161/auto.27442
Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, Rusten TE, Stenmark H, Brech A (2008) Ref(2)P, the drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol 180:6. https://doi.org/10.1083/jcb.200711108
Owusu-Ansah E, Banerjee U (2009) Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 461:7263. https://doi.org/10.1038/nature08313
Palmer WH, Dittmar M, Gordesky-Gold B, Hofmann J, Cherry S (2020) Drosophila melanogaster as a model for arbovirus infection of adult salivary glands. Virology 543:1–6. https://doi.org/10.1016/j.virol.2020.01.010
Papp D, Kovács T, Billes V, Varga M, Tarnóci A, Hackler L, Puskás LG, Liliom H, Tárnok K, Schlett K, Borsy A, Pádár Z, Kovács AL, Hegedűs K, Juhász G, Komlós M, Erdős A, Gulyás B, Vellai T (2016) AUTEN-67, an autophagy-enhancing drug candidate with potent antiaging and neuroprotective effects. Autophagy 12:2. https://doi.org/10.1080/15548627.2015.1082023
Parzych KR, Klionsky DJ (2014) An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 20:3. https://doi.org/10.1089/ars.2013.5371
Scott RC, Schuldiner O, Neufeld TP (2004) Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell 7(2):167–178. https://doi.org/10.1016/j.devcel.2004.07.009
Serbus LR, White PM, Silva JP, Rabe A, Teixeira L, Albertson R, Sullivan W (2015) The impact of host diet on wolbachia titer in Drosophila. PLoS Pathog 11:3. https://doi.org/10.1371/journal.ppat.1004777
Szatmári Z, Kis V, Lippai M, Hegedus K, Faragó T, Lorincz P, Tanaka T, Juhász G, Sass M (2014) Rab11 facilitates cross-talk between autophagy and endosomal pathway through regulation of hook localization. Mol Biol Cell 25:4. https://doi.org/10.1091/mbc.E13-10-0574
Takáts S, Pircs K, Nagy P, Varga Á, Kárpáti M, Hegedus K, Kramer H, Kovács AL, Sass M, Juhász G (2014) Interaction of the HOPS complex with syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell 25:8. https://doi.org/10.1091/mbc.E13-08-0449
Tang H, Ross A, Capel B (2008) Expression and functional analysis of Gm114, a putative mammalian ortholog of Drosophila bam. Deve biol 318(1):73–81
Teixeira F, Sanchez C, Hurd T, et al (2015) ATP synthase promotes germ cell differentiation independent of oxidative phosphorylation. Nat Cell Biol 17:689–696. https://doi.org/10.1038/ncb3165
Tracy K, Baehrecke EH (2013) The role of autophagy in Drosophila metamorphosis. Curr Top Dev Biol. https://doi.org/10.1016/B978-0-12-385979-2.00004-6
Wang SC, Hsu HJ, Lin GW, Wang TF, Chang CC, Lin M (2015) Germ plasm localisation of the HELICc of vasa in Drosophila: analysis of domain sufficiency and amino acids critical for localisation. Sci Rep. https://doi.org/10.1038/srep14703
White-Cooper H (2012) Tissue, cell type and stage-specific ectopic gene expression and RNAi induction in the Drosophila testis. Spermatogenesis 2:1. https://doi.org/10.4161/spmg.19088
Wucherpfennig T, Wilsch-Bräuninger M, González-Gaitán M (2003) Role of Drosophila Rab5 during endosomal trafficking at the synapse and evoked neurotransmitter release. J Cell Biol 161:3. https://doi.org/10.1083/jcb.200211087
Wullschleger S, Loewith R, Hall MN (2006) TOR signaling in growth and metabolism. Cell 124:3. https://doi.org/10.1016/j.cell.2006.01.016
Yang P, Zhang H (2014) You are what you eat: multifaceted functions of autophagy during celegans development. Cell Res 24:1. https://doi.org/10.1038/cr.2013.154
Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ (2010) Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465:7300. https://doi.org/10.1038/nature09076
Zhao S, Fortier TM, Baehrecke EH (2018) Autophagy Promotes Tumor-like Stem cell niche occupancy. Curr Biol 28:19. https://doi.org/10.1016/j.cub.2018.07.075
Acknowledgements
Drosophila stocks was obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537). Other Drosophila strains and reagents were kindly provided by Gábor Juhász (Department of Anatomy, Cell- and Developmental Biology, Eötvös Loránd University, Budapest, Hungary), Szabolcs Takáts (Department of Anatomy, Cell- and Developmental Biology, Eötvös Loránd University, Budapest, Hungary) and Rita Sinka (Department of Genetics, University of Szeged, Szeged, Hungary). The authors also thank Regina Preisinger, Beatrix Supauer and Erzsébet Gatyás for the excellent technical assistance.
Funding
Open access funding provided by Eötvös Loránd University. This work was supported by the OTKA grants (Hungarian Scientific Research Fund) K109349 and K132439 to TV, MEDinPROT Protein Science Research Synergy Program (provided by the Hungarian Academy of Sciences; HAS) to TV, the European Union and the State of Hungary, co-financed by the European Regional Development Found (VEKOP-2.3.2–16-2017–00014) to TV. TV was supported by the ELKH/MTA-ELTE Genetics Research Group (01062). This work was completed in the ELTE Institutional Excellence Program supported by The National Research, Development and Innovation Office (NKFIH-1157–8/2019-DT). TK was supported by the Bolyai Reseach Scholarship and the ÚNKP-20–5 New National Exellence Program Of The Ministry For Innovation And Technology Form The Source Of The National Research, Development And Innovation Fund.
Author information
Authors and Affiliations
Contributions
T.K. designed and performed experiments on Drosophila samples, analyzed data and wrote the manuscript; V.B.V. performed experiments on Drosophila samples, analyzed data and wrote the manuscript; D.S., F.Sz.. performed experiments on Drosophila samples, analyzed data; G.P. performed experiments on electron microscopy samples; and T.V. designed experiments, analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
We have no competing interests.
Ethical Approval
Research humans and non-human animals must include a statement detailing ethical approval and informed consent. Research must adhere to national and international guidelines and state that appropriate ethical approval and licences were obtained (Please refer to the number of your ethical permission).
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Varga, V.B., Schuller, D., Szikszai, F. et al. Autophagy is required for spermatogonial differentiation in the Drosophila testis. BIOLOGIA FUTURA 73, 187–204 (2022). https://doi.org/10.1007/s42977-022-00122-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42977-022-00122-7