Abstract
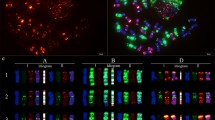
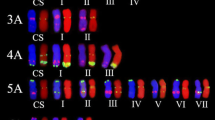
Although fluorescence in situ hybridization (FISH) karyotypes of wheat chromosomes have already been widely investigated, more probes are needed to enrich them. Novel karyotype characteristics of 166 common wheat cultivars bred from the hometown of Chinese Spring were revealed using 11 oligonucleotide (oligo) probes derived from nine kinds of tandem repeats. Chromosomes 5A, 3B and 1D showed the highest number (24, 18 and 6) of karyotype variations in A-, B-, and D-genome chromosomes, respectively. According to the FISH karyotypes, the analyzed wheat cultivars were divided into eight clusters that correspond to breeding entities. Thirty and 38 cultivars contain wheat-Haynaldia villosa 6AL.6VS and wheat-rye 1BL.1RS translocation chromosomes, respectively, indicating their high breeding value in Sichuan, China. Additionally, four and three types of 6AL.6VS and 1BL.1RS translocations were observed, respectively. Scarce translocations were detected. 'Neimai' series cultivars contain reciprocal translocations 5AS.3BS/5AL.3BL. Three cultivars contain reciprocal translocations 5BL.5BS-7BL/7BS.7BL-5BS that occurred at high frequency in European wheat cultivars, indicating that the translocation is also adapted to Sichuan environments. These results indicate that some special karyotypes exist in Sichuan wheat cultivars, showing the relationship between karyotype and adaptation. Richer FISH bands of wheat chromosomes were revealed, and this can be used to investigate meiotic recombination conveniently.




Similar content being viewed by others
Availability of data and material
The materials can be obtained by requiring.
Code availability
Not applicable.
References
Badaeva ED, Dedkova OS, Gay G, Pukhalskyi VA, Zelenin AV, Bernard S, Bernard M (2007) Chromosomal rearrangements in wheat: their types and distribution. Genome 50:907–926
Carvalho A, Guedes-Pinto H, Lima-Brito J (2013) Polymorphism of the simple sequence repeat (AAC)5 in the nucleolar chromosomes of old Portuguese wheat cultivars. J Genet 92:583–586
Cuadrado Á, Jouve N (2010) Chromosomal detection of simple sequence repeats (SSRs) using nondenaturing FISH (ND-FISH). Chromosoma 119:495–503
Friebe B, Gill BS (1994) C-band polymorphism and structural rearrangements detected in common wheat (Triticum aestivum). Euphytica 78:1–5
Guo J, Gao D, Gong W, Li H, Li J, Li G, Song J, Liu J, Yang Z, Liu C (2019) Genetic diversity in common wheat lines revealed by fluorescence in situ hybridization. Plant Syst Evol 305:247
Han FP, Lamb JC, Birchler A (2006) High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci USA 103:3238–3243
Huang X, Zhu M, Zhuang L, Zhang S, Wang J, Chen X, Wang D, Chen J, Bao Y, Guo J, Zhang J, Feng Y, Chu C, Du P, Qi Z, Wang H, Chen P (2018) Structural chromosome rearrangements and polymorphisms identified in Chinese wheat cultivars by high-resolution multiplex oligonucleotide FISH. Theor Appl Genet 131:1967–1986
Jiang M, Xaio ZQ, Fu SL, Tang ZX (2017) FISH karyotype of 85 common wheat cultivars/lines displayed by ND-FISH using oligonucleotide probes. Cereal Res Commun 45:549–563
Komuro S, Endo R, Shikata K, Kato A (2013) Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56:131–137
Liu D, Zhang L, Hao M, Ning S, Yuan Z, Dai S, Huang L, Wu B, Yan Z, Lan X, Zheng Y (2018) Wheat breeding in the hometown of Chinese spring. Crop J 6:82–90
McIntyre CL, Pereira S, Moran LB, Appels R (1990) New Secale cereale (rye) DNA derivatives for the detection of rye chromosome segments in wheat. Genome 33:635–640
Nagaki K, Tsujimoto H, Isono K, Sasakuma T (1995) Molecular characterization of a tandem repeat, Afa family, and its distribution among Triticeae. Genome 38:479–486
Paradis E, Schliep K (2018) Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35:526–528
Pollak Y, Zelinger E, Raskina O (2018) Repetitive DNA in the architecture, repatterning, and diversification of the genome of Aegilops speltoides Tausch (Poaceae, Triticeae). Front Plant Sci 9:1779
Ren T, He M, Sun Z, Tan F, Luo P, Tang Z, Fu S, Yan B, Ren Z, Li Z (2019) The polymorphisms of oligonucleotide probes in wheat cultivars determined by ND-FISH. Molecules 24:1126
Schneider A, Linc G, Molnár-Láng M (2003) Fluorescence in situ hybridization polymorphism using two repetitive DNA clones in different cultivars of wheat. Plant Breed 122:396–400
Sharma SK, Yamamoto M, Mukai Y (2016) Molecular cytogenetic approaches in exploration of important chromosomal landmarks in plants. In: Rajpal R, Rao SR, Raina SN (eds) Sustainable development and biodiversity. Springer Press, Geneva, pp 127–148
Tang Z, Yang Z, Fu S (2014) Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genet 55:313–318
Tang S, Qiu L, Xiao Z, Fu S, Tang Z (2016) New Oligonucleotide probes for ND-FISH analysis to identify barley chromosomes and to investigate polymorphisms of wheat chromosomes. Genes 7:118
Tang S, Tang Z, Qiu L, Yang Z, Li G, Lang T, Zhu W, Zhang J, Fu S (2018) Developing new oligo probes to distinguish specific chromosomal segments and the A, B, D genomes of wheat (Triticum aestivum L.) using ND-FISH. Front Plant Sci 9:1104
Walkowiak S, Gao L, Monat C, Haberer G, Kassa MT, Brinton J et al (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature 588:277–283
Xiao Z, Tang S, Qiu L, Tang Z, Fu S (2017) Oligonucleotides and ND-FISH displaying different arrangements of tandem repeats and identification of Dasypyrum villosum chromosomes in wheat backgrounds. Molecules 22:973
Yang M, Yang Z, Yang W, Yang E (2018) Development and identification of new wheat varieties (lines) with multiple translocation chromosomes via cytogenetic methods. J Triticeae Crops 38:127–133
Zou Y, Wan L, Luo J, Tang Z, Fu S (2021) FISH landmarks reflecting meiotic recombination and structural alterations of chromosomes in wheat (Triticum aestivum L.). BMC Plant Biol 21:167
Acknowledgements
We are thankful to Ennian Yang, Ling Wu, Zongjun Pu (Crop Research Institute, Sichuan Academy of Agricultural Sciences, China;), Yong Ren (Mianyang Branch of National Wheat Improvement Center, Mianyang Institute of Agricultural Sciences, China), Tao Wang (Chengdu Institute of Biology, Chinese Academy of Sciences, Chengdu, China) and Suizhuang Yang (School of Life Science and Engineering, Southwest University of Science and Technology, China) for their kindly providing seeds. This manuscript is provided by the National Natural Science Foundation of China (No. 32070373), the Science and Technology Project of Sichuan, China (No. 2020YJ0128) and the “13th Five-Year” Crops, Livestock and Poultry Breeding Program of Sichuan Province (No. 2016NY0030).
Funding
This manuscript is provided by the National Natural Science Foundation of China (No. 32070373), the Science and Technology Project of Sichuan, China (No. 2020YJ0128) and the “13th Five-Year” Crops, Livestock and Poultry Breeding Program of Sichuan Province (No. 2016NY0030).
Author information
Authors and Affiliations
Contributions
SF, ZT, MH, and DL conceived and designed the study. ZT, JL and MH collected materials. ZH, JL, LW and JL performed the experiments. ZH, JL, LW and MH analyzed data. ZT wrote original draft preparation. SF, MH, and DL reviewed and edited this manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
This research has no conflict of interest.
Ethics approval
Not applicable.
Additional information
Communicated by M. Molnár-Láng.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hu, Z., Luo, J., Wan, L. et al. Chromosomes polymorphisms of Sichuan wheat cultivars displayed by ND-FISH landmarks. CEREAL RESEARCH COMMUNICATIONS 50, 253–262 (2022). https://doi.org/10.1007/s42976-021-00173-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42976-021-00173-x