Abstract
Food web research needs to be predictive in order to support decisions system-based conservation. In order to increase predictability and applicability, complexity needs to be managed in such a way that we are able to provide simple and clear results. One question emerging frequently is whether certain perturbations (environmental effects or human impact) have positive or negative effects on natural ecosystems or their particular components. Yet, most of food web studies do not consider the sign of effects. Here, we study 6 versions of the Kelian River (Borneo) food web, representing six study sites along the river. For each network, we study the signs of the effects of a perturbed trophic group i on each other j groups. We compare the outcome of the relatively complicated dynamical simulation model and the relatively simple loop analysis model. We compare these results for the 6 sites and also the 14 trophic groups. Finally, we see if sign-agreement and sign-determinacy depend on certain structural features (node centrality, interaction strength). We found major differences between different modelling scenarios, with herbivore-detritivore fish behaving in the most consistent, while algae and particulate organic matter behaving in the least consistent way. We also found higher agreement between the signs of predictions for trophic groups at higher trophic levels in sites 1–3 and at lower trophic levels in site 4–6. This means that the behaviour of predators in the more natural sections of the river and that of producers at the more human-impacted sections are more consistently predicted. This suggests to be more careful with the less consistently predictable trophic groups in conservation management.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
In complex ecological systems, the multiplicity of direct and indirect interactions makes it difficult to provide simple and clear predictions on the effect of single-node perturbations. The effects on other organisms and, generally, community response are the outcome of a number of interconnected pathways. Predicting whether the influence of organism i on organism j will be positive or negative is not easy, even without considering non-trophic effects and complicated functional responses. Also, the experimental results on positive inter-specific interactions are still quite sporadic (Bertness & Shumway, 1993; Bruno et al., 2003; Kareiva & Bertness, 1997) compared to the mass of literature on competition and predation. Considering effect sign is critically important if food web models are to be extended to ecological interaction networks: this is the way how to complete antagonistic predator–prey interactions with positive feedback loops (Dong et al., 2020; Ulanowicz, 1995) and mutualisms (Leemans et al., 2020).
Network analysis offers various methods for predicting effect sign. Loop analysis (Levins, 1974; Puccia & Levins, 1985a), mixed trophic impact (Bondavalli & Ulanowicz, 1999; Ulanowicz & Puccia, 1990), signed topological importance (Liu et al., 2010, 2020) and dynamical simulations (Jordán et al., 2012) can be used but their relationship is not straightforward (i.e. whether they provide similar or complementary information). Yet, loop analysis has been used extensively together with topological network analysis (e.g. Ortiz et al., 2013, 2015, 2017).
Generally speaking, topological models provide fast and easy but not very realistic results: they consider only the existence of network links (in a binary way) or also their directionality and the weights on them (in more detailed network models) but do not reflect the dynamical behaviour (parameters, kinetics) of the network. Sophisticated food web simulations may provide more accurate results but typically need a lot of data (dynamical parameters) and the interpretation of results raises questions about indeterminacy (Yodzis, 1988), uncertainty (Geary et al., 2020), additivity (Móréh et al., 2021) or whether the system is close to equilibrium (Baker et al., 2017). Qualitative models, like loop analysis, are somewhere in between, trying to combine simplicity (without explicit dynamics) and reality (interaction sign considered).
In this paper, we study food webs at the level of individual nodes (trophic groups), at the level of the interactions among the trophic groups and at the level of the whole networks. We compare (1) the results of loop analysis and dynamical simulations predicting the sign of effects following single-node perturbations. Beyond comparing the signs of the predictions in these two methods, we also compare (2) the 6 food web models as well as (3) the 14 trophic groups. Finally, (4) we investigate if there is a correlation between the network centrality of a trophic group and the sign-agreement between the two models predicting its effects on other groups.
Data and methods
Data
We used 6 versions of the Kelian River (Borneo) food web for our analysis (data from Yule, 1995; Yule et al., 2010). This makes it possible to assess spatial variability within the ecosystem.
The food web is described for 6 different locations along the river (Jordán et al., 2012, 2017), based on extensive earlier field work (Yule, 1995; Yule et al., 2010). The 6 sites represent a gradient from a pristine rainforest to a human settlement. The pristine food web (site 1) contains 14 trophic groups and 2 additional groups appear only downstream, so the total number of trophic groups is 16 (see Table 1). Most of the groups represent living organisms (e.g. PRED: invertebrate predators) but there are some non-living groups as well (e.g. POM: settled and suspended coarse and fine Particulate Organic Matter). The definition of the groups and the description of interactions among them are based on long-term, extensive field work (Yule, 1995; Yule et al., 2010).
Food web structure: topological importance
We quantified the network centrality of trophic groups by the topological importance index (TI). This method is based on an earlier measure (Müller et al., 1999) for 2-step-long apparent competition in host-parasitoid communities, later generalized for n-step-long indirect effects in Jordán et al. (2003). Consider that i and j are connected (prey and predator), and the direct effect of i on j (aij) is as follows:
where Dj is the degree of node j (the number of direct neighbours). So, if i is the only neighbour of j, its effect will equal 1 (the maximum), but if j has more neighbours, the effect of i will be smaller. We can put this direct effect between all pairs of nodes in a matrix A, and generalize it to an n-steps effect just by calculating An. As different paths of different lengths between two nodes may exist, we can calculate the effects of node i on j, up to a defined number of steps, and then average them over the maximum number of steps considered (i.e. n):
with this average effect of all pairs in the network, we can construct an interaction matrix IMn, where the ijth element is the AEn,ij. Then, the sum of the values in ith row is the topological importance of i, as it is the sum of effects up to n-steps on the other nodes of the network.
Based on indirect chain effects, we can assume that central species of interaction networks are of higher importance for the rest of the community (Jordán et al., 2006). This is one approach to the quantification of key (possibly keystone) species in food webs. The centrality of species (trophic groups) in food webs seems to be a systemic property with important ecological correlates (e.g. body size, mobility, see: Olmo Gilabert et al., 2019). There are a number of topological indices quantifying centrality in networks and their relationship is increasingly understood (Scotti et al., 2007). Since we must deal with indirect effects in order to assess the combinations of positive and negative impacts, we use the TI index, explicitly considering the length of direct and indirect pathways in food webs (Jordán et al., 2003). We note that TI is an useful centrality measure out of the many, according to a recent study where machine learning identified the most predictive combinations of centrality indices (Gouveia et al., 2021). In this paper, we used TI3, i.e. we considered indirect effects up to 3 steps. We performed these calculations in order to see whether there is structural basis of (constraints on) the sign-agreement (for nodes and networks) and sign-determinacy (for interactions and networks).
Loop analysis
Loop analysis is a qualitative method that uses signed digraphs to illustrate networks of interacting variables (Levins, 1974; Puccia & Levins, 1985a). This technique gives the opportunity to represent the structure of linkages of the variables and the patterns of their variations (Bodini & Clerici, 2016; Dambacher & Ramos-Jiliberto, 2007). Also, these models allow to examine the effect of non-biological variables on our system (such as gold mining, fishing etc.). This qualitative modelling approach provides a method that is useful where species and their natural history are well-known, but not quantified (Dambacher et al., 2003).
Signed digraphs are based on the generally accepted interactions between nodes (trophic groups). These interactions come from the previous trophic models in Jordán et al. (2012). The figures show two types of connections: arrows (→) for positive and circle-head links (−o) for negative effects. These links are originated from the coefficients of the community matrix (Levins, 1974). The diagonal terms of the community matrices are self-effects on system variables, represented in signed digraphs as links connecting variables with themselves. These links are self-dampening (circle-headed) with self-limiting growth rate. For a simple prey–predator system, see Fig. 1.
Using the qualitative modelling framework of loop analysis, one can analyse pathways and feedbacks in the system, making predictions about the response of variables to perturbations. These can be the addition (increased biomass) or deletion (decreased biomass) of other nodes. Based on feedbacks and pathways, one can qualitatively specify the direction of changes.
We followed the method described in Bodini and Clerici (2016) to get the predictions for our networks. The loop formula is used for calculating the equilibrium value of the variables following a perturbation, so it can be deduced how does the abundance of a certain variable change (Bodini, 2000):
On the left side, xj is the variable with the equilibrium value being calculated and c is the changing parameter (e.g. mortality, fecundity, abundance). On the right side, f is the growth rate, ∂fi/∂c designates whether the growth rate of the ith variable is increasing or decreasing (positive or negative input, respectively), pji(k) is the pathway connecting the variable to the changed biomass variable (where the perturbation enters the system), Fn-k(comp) is the complementary feedback, which buffers or reverses the effects of the pathway and Fn designates the overall feedback of the system, which is a measure of the inertia of the whole system to change (Bodini, 2000; Bodini & Clerici, 2016). See also (Puccia & Levins, 1985a) for the discussion of the correspondence between matrix algebra and loop analysis.
The net effect (the sum of the direct and indirect effects) on variable i resulting from a perturbation on variable j is given by the j − ith element of the inverse community matrix [A]−1 (see Levins, 1974; Puccia & Levins, 1985b; Raymond et al., 2011). The sign of the coefficients of [A]−1 gives the direction of the expected changes for the variables (Bodini & Clerici, 2016). To make predictions, we used a routine that randomly assigns numerical values from an uniform distribution to the coefficients of the community matrix (these coefficients belongs to the links of the signed digraph). This was performed 100 * N2 times, where N is the number of variables in the system. Matrices satisfying the asymptotic Lyapunov criteria were accepted and inverted. The routine of Bodini and Clerici (2016) calculated predictions for the probabilities based on the percentage of positive and negative signs and zeroes in the inverted matrices. They defined a set of rules to make a final table of predictions only from signs (Table 2, Bodini & Clerici, 2016).
Community matrices show the links between the trophic groups. The elements in the rows affect the elements in the columns and the values could be − 1, 0 or 1. These represent prey–predator (resource-consumer) effects. We decided to work with self-dampening variables only for living groups (see Table 3 for the community matrix of site 1).
We were interested in the effect of decreasing the biomass of each trophic group (i.e. the value of each variable), one by one. For this, we needed to simply reverse our predictions (change signs from + to −) (Dambacher et al., 2002). By predicting these changes, qualitative models can also predict the correlation patterns between the examined groups/variables (Bodini, 2000; Bodini & Clerici, 2016; Puccia & Levins, 1985a).
Levins’ loop algorithm was extended in Dambacher et al. (2002) to complex ecological systems. The adjoint of the negative of this community matrix shows the net number of complementary feedback cycles. A complementary feedback appears if k variables in the path are ideally excluded from the graph: what remains is called the complementary subsystem. The complementary feedback is the feedback that groups all the variables in the complementary subsystem (Bodini & Clerici, 2016) that contribute to the responses of other variables in the whole system. Therefore, the adjoint of the -°A is equivalent to Levins’ loop analysis algorithm and its relation with the inverse matrix (A−1) is as follows:
where the adjoint and inverse matrices are calculated with the negative of the community matrix, thus the positive input is read down the columns and the responses along the rows (Dambacher et al., 2002). The adjoint quantifies the results of the prediction tables, so their numerical values make correlation tests possible in further analyses.
An “absolute feedback” matrix was defined in Dambacher et al. (2002) to calculate the absolute number of complementary feedback cycles in a response whether positive or negative:
where ˙A denotes the adjacency matrix (absolute values of °A). Complementary feedback cycles are needed to express the equilibrium response of community members by inverting the direct effects of the community matrix (Dambacher et al., 2003).
The “weighted predictions” matrix can be calculated from Eqs. (1)–(2):
where “→” is a vectorized matrix operator, what denotes element-by-element division (Wij = 1, when Tij = 0). The elements of W show the probability of sign-determinacy of response predictions in the adjoint matrix. If all of them are of the same sign in one cell, then Wij = 1. If there is an equal number of negative and positive feedback cycles, then Wij = 0 (Dambacher et al., 2002). The Wij = 0.5 value is a threshold for sign-determinacy in models of any size (Dambacher et al., 2003).
We used the adjoint values to investigate if sign-determinacy depends on the strength of interactions (based on both structure and simulations).
Dynamical simulations
For the dynamical simulation data on the food webs, we used earlier results of an individual-based model (Jordán et al., 2012, 2017). This simulation model was built in the process algebra-based language, BlenX (Jordán et al., 2011): abundance was calculated for the trophic groups and interaction strength was converted to probability, according to the kinetics of the stochastic simulation framework used routinely in systems biology (Dematté et al., 2008; Gillespie, 1977; Priami, 2009; Priami & Quaglia, 2004).
The stochastic IBM simulation model was parameterized with field data (e.g. abundance data, Yule, 1995; Yule et al., 2010) and was balanced by a genetic algorithm. Following the reference runs, sensitivity analysis was performed. The abundance of each trophic group was perturbed, and the response of each other trophic group was measured. Since this was a stochastic model, both the mean and the variability were evaluated (for the details, see Jordán et al., 2012, 2017).
In this paper, we used only the sign of the responses (not their strength). For comparability, minimal changes must be made in the results of the dynamical simulations: since these categories do not exist in simulations, ? + , ? − and 0* predictions were considered as + , − and 0, respectively. Further, as the effects in dynamical simulations never result exactly in 0, we needed to define zero effects during the simulations. For this, we needed a threshold level below the effect can be considered zero. Instead of using an arbitrary threshold, we decided to set the threshold in such a way that the number of zeroes equals the number of zero responses in loop analysis. Values in this “corridor” of the smallest positive and negative simulation outcomes were considered 0.
Software
We used MASS 7.3–51.5 and nlme 3.1–148 R packages for the analyses. For simulating sign predictions, we used the R code in Bodini and Clerici (2016). The LoopAnalyst 1.2–6 package was used for the calculations of the adjoint of the -°A and the “weighted predictions” matrices (and it required the nlme package, too). Figures 2 and 3 are created with GVEdit Graph File Editor For Graphviz version: 1.02 and Graphviz version:2.38.
Statistical methods
Sign-agreement was examined on several levels. We compared the predictions of loop analysis to the calculations made by the structural importance index TI3 and to the results of dynamical simulations for the whole networks, for individual nodes (i.e. the rows of the matrices) and for individual interactions.
As described above, we categorized the mean outcome of dynamical simulation results into 3 categories (+ , 0, −). This way, the table of predictions from loop analysis and the matrix of simulation effects became comparable (both containing only + , − and 0). A binary sign-agreement matrix contained 1 (yes) for similar and 0 (no) for different signs in the two matrices. The percentage of 1 values served for quantifying sign-agreement (1 s in the main diagonal, corresponding to self-effects, were not considered).
Chi-square tests were applied for the effect signs for each node, determining if there is significant difference in either loop analysis or dynamical simulations.
In order to see if there is some structural basis for sign-determinacy and sign-agreement, we tested the correlation between (1) the adjoint matrix and the dynamic simulation results at the level of the whole network, (2) the adjoint matrix and interaction strength calculated by TIij at the level of interactions, (3) the dynamic simulation results and interaction strength calculated by TIij at the level of interactions and (4) sign prediction of loop analysis and node centrality measured by TIi at the level of network nodes.
Results
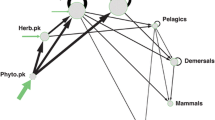
Figure 2 shows the food web for each of the 6 sites. The number of trophic groups varies between 12 and 15, starting from 14 in the pristine forest (site 1).
Our values in the “weighted predictions” matrix were mostly under Wij = 0.5. According to this, most of the prediction signs were indeterminate. The predicted signs of effects (both clear and weaker predictions) for each trophic group are shown in Fig. 3, for each of the 6 sites (Fig. 3a–f). In site 1 (Fig. 3a), mutually negative (CARN-HEDE), mutually positive (CARN-OMNI), antagonistic (LEAF-GRAZ) uni-directional positive (TERR-PRED) and uni-directional negative (HERB-CARN) interactions can be seen. Between some pairs of trophic groups, there is no interaction predicted (e.g. CARN and COLG). Higher sign-agreement is seen for organisms at higher trophic levels. This pattern seems to be the same also for site 2 (Fig. 3b) but not for the other sites. Site 4 (Fig. 3d) is simpler than the previous sites, with some groups having only a single interaction partner (e.g. CARN, HEDE, COLG). The differences among the sites are quite characteristic in loop analysis, while the simulation models vary less. Site 6 (Fig. 3f) is strongly dominated by factors essentially external to the stream ecosystem (TERR and HUMW).
Food web of the Kelian River ecosystem in sites 1–6 (a–f). Arrows show the predictions of loop analysis: the sign of the effect of decreasing the abundance of a node on another (red is positive, blue is negative). The colour of nodes shows sign-agreement (%) between the predictions of loop analysis and the results of dynamical simulations (increasing from light yellow to dark green). Whether perturbing the light yellow nodes has positive or negative effect on others largely depends on the model chosen (see Table 4). On the contrary, the perturbation of dark green nodes is similarly predicted by both approaches (see Table 4). Coloured by ColorBrewer (Neuwirth & Brewer, 2014)
Sign-agreement predicted by loop analysis and by the dynamic simulations are shown in Table 4, at the level of the whole networks and for individual nodes (this latter is represented by the colours in Fig. 3: sign-agreement increases from light yellow to dark green). The mean correspondence of signs at the network level is under 50%. Maximum sign-agreement ranged between 50 and 64.29%, and the minimum was between 7.14 and 27.27% (see Table 4). On average, site 6 in the Kelian River is the ecosystem where loop analysis best predicts the signs of dynamical simulation results. From site 2 to site 6, there is a monotonous increase in mean (and the minimal) sign-agreement (the maximum values show no trend and also site 1 is out of this pattern). This suggests that the two methods provide different information on the ecosystem. Some trophic groups generally give consistent behaviour (e.g. HEDE), while others (ALGA, POM) behave differently in different modelling environments. Whether POM will have positive or negative impact on others is differently predicted in 5 out of 6 sites.
Sign-agreement was measured also at the level of the interactions between trophic groups. The mean of sign-agreement is also under 50% but the maximums are between 50 and 72% (Table 5). The high sign-agreement for HEDE is seen also in the interactions in this group, being involved in 4 out of 7 of the highest-agreement interactions. It is also noted that the same group (HEDE) can be involved in minimum (HEDE-POM) and maximum (HEDE-TERR) agreement interactions (see site 2). It is noted that COLG in site 5 has several minimal-agreement interactions (e.g. COLG-POM), after being the most predictable group in site 4 (with the most predictable interaction, COLG-CARN).
The Chi-square tests showed that the predictions of loop analysis for different trophic groups were significantly different only for site 4 of Kelian River (Fig. 4a, see Supplementary 1 for all of the other simulations), while the Chi-square tests for the results of dynamic simulations did not show significant differences (Fig. 4b). Based on loop analysis (Fig. 4a), CARN, COLG and HEDE have predominantly zero and occasionally positive effects on others. On the contrary, TERR (and to some extent POM) have the highest proportion of negative effects. Dynamical simulations (Fig. 4b) show a different outcome with DIAT being the only one without negative effects and HEDE (and ALGA) with the largest negative impacts. According to Supplementary 1, there are significant differences among the distribution of impact signs of various organisms in almost no cases. In some cases, this homogeneity is quite strong (e.g. site 3, loop analysis with 3 categories), while there are models with non-significant but visible differences (e.g. GRAZ and HEDE in the simulation model of site 2). In the model of site 4, loop analysis with 5 sign categories also provided significant differences among the organisms: the pattern is qualitatively similar to the result of the model with 3 sign categories (see Fig. 4). Only the structural interaction strength (TIij) and the adjoint values (LAadj) showed significant difference among the various trophic groups in site 4 of the model (see Supplementary 1).
The number of positive (green), negative (red) and zero (blue) effects. Each 14 functional group are listed along the x axis, and the 13 other are counted along the y axis. This figure represents site 4 of the Kelian River ecosystem model, based on loop analysis (a) and dynamical simulations (b). In order to make them comparable, the predictions of loop analysis were transformed: we used only 3 categories, + ? was replaced by + , − ? was replaced by − and 0* was replaced by 0. The sign composition of organisms differs significantly in a (χ222 = 34,67; p = 0.04193) but not in b (χ222 = 22,989; p = 0.4024). The same plots for other ecosystem models as well as the plots for loop analysis with 6 prediction categories are shown in Supplementary 1
Spearman correlation (ǀρǀ values) between pairs of simulated interaction strength (aij), interaction strength based on food web topology (TIij), node centrality (TIi), sign-agreement based on loop analysis predictions (LApred) and the adjoint value based on loop analysis (LAadj) revealed only a single significant correlation (Supplementary 2). In site 4, simulated interaction strength (aij) correlated with the adjoint (LAadj).
Conclusion
Predictive food web research would be a quantitative and holistic toolkit for systems-based conservation efforts. Good predictions on strong and weak as well as positive and negative effects are important for management and policy. While dynamical simulation exercises require a large number of parameters and complicated models, the semi-quantitative methodology of loop analysis offers simpler (only effect sign) and faster (no temporal simulations) results, generally easier to understand and interpret (under the conditions of close-to-equilibrium situations). Topological analyses are the simplest and least realistic ones. The major question was to what extent and when simpler methods can replace the more complicated ones. We show that for some organisms in some locations (higher trophic levels in more pristine river segments and lower trophic levels in the more human-impacted river segments), the predictions of the different models are in greater agreement. For these cases, one can trust more the model results. In the opposite cases (lower trophic levels in the more pristine river segments and higher trophic levels in the more human-impacted river segments), the predictions are more different, so model results must be more carefully accepted. In these latter cases, more research, better data or a different modelling framework may be needed in order to better support conservation management.
Comparing the sites, we see that the relatively consistent predictability of herbivore-detritivore fish (HEDE) is seen in the sites of otherwise contrasting signs (site 2, site 3). In the middle of the food web, their dynamics may not be extremely affected, or unexpected dynamical effects may be balanced by multiple interactions. The most consistent interaction happens between terrestrial insects (TERR) and carnivorous fish (CARN) in the most consistent model of site 6. Here, the strong bottom-up dominance in regulation (human waste detritus) simplifies community dynamics but the external input of terrestrial insects remains consumed by carnivorous fish independently of this bottom-up dominance. This is the case when loop analysis and the dynamical simulation model mostly agree on sign prediction.
Based on Table 4, the general pattern emerges that higher sign-agreement is characteristic for trophic groups at higher trophic levels in sites 1–3, while trophic groups at lower trophic levels in sites 4–6.
Supposing that both models are relevant and correct, the question emerges why and to what extent should they be similar to each other. Similarity means that one reinforces the other, while differences may suggest that they are complementary, providing different kind of information. In the future, mixed trophic impact analysis, signed topological importance and further kinds of dynamical simulations must also be compared to the models studied here. If the predictions of different models agree, one can trust better them, while if results vary, more research or better data are needed. Furthermore, models explicitly addressing uncertainty (Rendall et al., 2021) and predictability (Adams et al., 2020) are becoming increasingly important for supporting conservation management.
It is a question if experimental studies (e.g. mesocosm experiments) could help to better understand the predictability of effect signs and their differences in different modelling environments. Performing experiments only for a few species (e.g. perturbing COLG) might help a lot in calibrating the amount of expected changes and scale the models.
In future research, we need to better understand how trophic complexity is related to predictability, in terms of either interaction strengths or interaction signs. Earlier research, based on time series data, found quite convincing results on the dimensionality of niche space (Stenseth et al., 1997). Also, our research can be extended, for example, towards (1) considering the asymmetry of interactions and (2) considering loop sign (Harary, 1959; Wey et al., 2019). The former could be important in order to better understand cause-effect relationships, as strong asymmetry may imply causality. The latter could help to better assess network-level stability.
Abbreviations
- ALGA:
-
Green and blue-green algae
- CARN:
-
Carnivorous fish
- COLF:
-
Invertebrate collector-filterers
- COLG:
-
Invertebrate collector-gatherers
- DIAT:
-
Diatoms
- FILA:
-
Filamentous bacteria
- GRAZ:
-
Invertebrate grazers
- HEDE:
-
Herbivore-detritivore fish
- HERB:
-
Herbivorous fish
- HUMW:
-
Human waste
- LEAF:
-
Leaf litter
- OMNI:
-
Omnivorous fish
- POM:
-
Settled and suspended coarse and fine Particulate Organic Matter
- PRED:
-
Invertebrate predators
- SHRE:
-
Invertebrate shredders
- TERR:
-
Terrestrial insects
References
Adams, M. P., Sisson, S. A., Helmstedt, K. J., Baker, C. M., Holden, M. H., Plein, M., Holloway, J., Mengersen, K. L., & McDonald-Madden, E. (2020). Informing management decisions for ecological networks, using dynamic models calibrated to noisy time-series data. Ecology Letters, 23, 607–619.
Baker, C. M., Gordon, A., & Bode, M. (2017). Ensemble ecosystem modeling for predicting ecosystem response to predator reintroduction. Conservation Biology, 31, 376–384.
Bertness, M. D., & Shumway, S. W. (1993). Competition and facilitation in marsh plants. American Naturalist., 142, 718–724.
Bodini, A. (2000). Reconstructing trophic interactions as a tool for understanding and managing ecosystems: Application to a shallow eutrophic lake. Canadian Journal of Fisheries and Aquatic Sciences, 57, 1999–2009.
Bodini, A., & Clerici, N. (2016). Vegetation, herbivores and fires in savanna ecosystems: A network perspective. Ecological Complexity, 28, 36–46.
Bondavalli, C., & Ulanowicz, R. E. (1999). Unexpected effects of predators upon their prey: The case of the American alligator. Ecosystems, 2, 49–63.
Bruno, J. F., Stachowitz, J. J., & Bertness, M. D. (2003). Inclusion of facilitation into ecological theory. Trends in Ecology and Evolution, 18, 119–125.
Dambacher, J. M., & Ramos-Jiliberto, R. (2007). Understanding and predicting effects of modified interactions through a qualitative analysis of community structure. The Quarterly Review of Biology, 82(3), 227–250.
Dambacher, J. M., Li, H. W., & Rossignol, P. A. (2002). Relevance of community structure in assessing indeterminacy of ecological predictions. Ecology, 83, 1372–1385.
Dambacher, J. M., Li, H. W., & Rossignol, P. A. (2003). Qualitative predictions in model ecosystems. Ecological Modelling, 161, 79–93.
Dematté, L., Priami, C., Romanel, A., & Soyer, O. (2008). Evolving BlenX programs to simulate the evolution of biological networks. Theoretical Computer Science, 408, 83–96.
Dong, X., Grimm, N. B., Heffernan, J. B., & Muneepeerakul, R. (2020). Interactions between physical template and self-organization shape plant dynamics in a stream ecosystem. Ecosystems, 23, 891–905.
Geary, W. L., Bode, M., Doherty, T. S., et al. (2020). A guide to ecosystem models and their environmental applications. Nature Ecology and Evolution, 4, 1459–1471.
Gillespie, D. T. (1977). Exact stochastic simulation of coupled chemical reactions. Journal of Physical Chemistry, 81, 2340–2361.
Gouveia, C., Móréh, Á., & Jordán, F. (2021). Combining centrality indices: maximizing the predictability of keystone species in food webs. Ecological Indicators, 126, 107617.
Harary, F. (1959). Status and contrastatus. Sociometry., 22, 23–43.
Jordán, F., Gjata, N., Mei, S., & Yule, C. M. (2012). Simulating food web dynamics along a gradient: Quantifying human influence. PLoS ONE, 7, e40280.
Jordán, F., Scotti, M., & Yule, C. M. (2017). Food web simulations: stochastic variability and systems-based conservation. In J. C. Moore, P. C. de Ruiter, K. S. McCann, & V. Wolters (Eds.), Adaptive food webs (pp. 342–351). Cambridge University Press.
Jordán, F., Liu, W. J., & Davis, A. J. (2006). Topological keystone species: Measures of positional importance in food webs. Oikos, 112, 535–546.
Jordán, F., Liu, W.-C., & van Veen, F. J. F. (2003). Quantifying the importance of species and their interactions in a host-parasitoid community. Community Ecology, 4, 79–88.
Jordán, F., Scotti, M., & Priami, C. (2011). Process algebra-based models in systems ecology. Ecological Complexity, 8, 357–363.
Kareiva, P. M., & Bertness, M. D. (1997). Re-examining the role of positive interactions in communities. Ecology, 78, 1945.
Leemans, L., Martínez, I., van der Heide, T., van Katwijk, M. M., & van Tussenbroek, B. I. (2020). A mutualism between unattached coralline algae and seagrasses prevents overgrazing by sea turtles. Ecosystems, 23, 1631–1642.
Levins, R. (1974). Qualitative analysis of partially specified systems. Annals New York Academy of Sciences, 231, 123–138.
Liu, W. C., Chen, H. W., Jordán, F., Lin, W. H., & Liu, W. J. (2010). Quantifying the interaction structure and the topological importance of species in food webs: A signed digraph approach. Journal of Theoretical Biology, 267, 355–362.
Liu, W. C., Huang, L. C., Liu, C. W., & Jordán, F. (2020). A simple approach for quantifying node centrality in signed and directed social networks. Applied Network Science, 5, 46.
Móréh, Á., Endrédi, A., Piross, I. S., & Jordán, F. (2021). Topology of additive pairwise effects in food webs. Ecological Modelling., 440, 109414.
Müller, C. B., Adriaanse, I. C. T., Belshaw, R., & Godfray, H. C. J. (1999). The structure of an aphid-parasitoid community. Journal of Animal Ecology, 68, 346–370.
Neuwirth, E., & Brewer, R. C. (2014). ColorBrewer palettes. R package version, 1.
Olmo Gilabert, R., Navia, A. F., De La Cruz-Agüero, G., Molinero, J. C., Sommer, U., & Scotti, M. (2019). Body size and mobility explain species centralities in the Gulf of California food web. Community Ecology., 20, 149–160.
Ortiz, M., Rodriguez-Zaragosa, F., Hermosillo-Nunez, B., & Jordán, F. (2015). Control strategy scenarios for the alien lionfish Pterois volitans in Chinchorro Bank (Mexican Caribbean) based on semi-quantitative loop network analysis. PLoS ONE, 10, e0130261.
Ortiz, M., Hermosillo-Nuñez, B., González, J., Rodríguez-Zaragoza, F., Gómez, I., & Jordán, F. (2017). Quantifying keystone species complexes: Ecosystem-based conservation management in the King George Island (Antarctic Peninsula). Ecological Indicators, 81, 453–460.
Ortiz, M., Levins, R., Campos, L., Berrios, F., Campos, F., Jordán, F., Hermosillo, B., Gonzalez, J., & Rodriguez, F. (2013). Identifying keystone trophic groups in benthic ecosystems: Implications for fisheries management. Ecological Indicators, 25, 133–140.
Priami, C. (2009). Algorithmic systems biology. Communications of the ACM, 52, 80–89.
Priami, C., & Quaglia, P. (2004). Modelling the dynamics of biosystems. Briefings in Bioinformatics, 5, 259–269.
Puccia, C. J., & Levins, R. (1985a). Qualitative modelling of complex systems: An introduction to loop analysis and time averaging. Harvard University Press.
Raymond, B., McInnes, J., Dambacher, J. M., Way, S., & Bergstrom, D. M. (2011). Qualitative modelling of invasive species eradication on subantarctic Macquarie Island. Journal of Applied Ecology, 48, 181–191.
Rendall, A. R., Sutherland, D. R., Baker, C. M., Raymond, B., Cooke, R., & White, J. G. (2021). Managing ecosystems in a sea of uncertainty: invasive species management and assisted colonizations. Ecological Applications, 31, e2306.
Scotti, M., Podani, J., & Jordán, F. (2007). Weighting, scale dependence and indirect effects in ecological networks: a comparative study. Ecological Complexity, 4(3), 148–159.
Stenseth, N. C., Falck, W., Bjørnstad, O. N., & Krebs, C. J. (1997). Population regulation in snowshoe hare and Canadian lynx: Asymmetric food web configurations between hare and lynx. Proceedings of the National Academy of Sciences, 94, 5147–5152.
Ulanowicz, R. E. (1995). Utricularia’s secret: The advantage of positive feedback in oligotrophic environments. Ecological Modelling, 79, 49–57.
Ulanowicz, R. E., & Puccia, C. J. (1990). Mixed trophic impacts in ecosystems. Coenoses, 5, 7–16.
Wey, T., Jordán, F., & Blumstein, D. (2019). Transitivity and structural balance in animal social networks. Behavioral Ecology and Sociobiology, 73, 88.
Yodzis, P. (1988). The indeterminacy of ecological interactions as perceived through perturbation experiments. Ecology, 69, 508–515.
Yule, C. M. (1995). The impact of sediment pollution on the benthic invertebrate fauna of the Kelian River, East Kalimantan Indonesia. Tropical Limnology, 3, 61–75.
Yule, C. M., Boyero, L., & Marchant, R. (2010). Effects of sediment pollution on food webs in a tropical river (Borneo, Indonesia). Marine and Freshwater Research, 61, 204–213.
Acknowledgements
I’m grateful to Catherine M. Yule for the data and earlier discussions on the database and to Sándor Imre Piross and István Reguly for earlier discussions on the methods.
Funding
Open access funding provided by ELKH Centre for Ecological Research. This output reflects only the author’s view and the European Union cannot be held responsible for any use of the information contained therein.
Author information
Authors and Affiliations
Contributions
V.F. performed the loop analysis modelling and the statistical analysis. She wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Supplementary Information
Below is the link to the electronic supplementary material.
42974_2021_68_MOESM1_ESM.xlsx
The number of positive (green), negative (red) and zero (blue) effects of each of the 12 functional groups (listed along the x axis) on each 11 others (counted along the y axis). Plots are based on loop analysis with 6 prediction categories, loop analysis with 3 prediction categories ( + ? replaced by +, − ? replaced by −, 0 * replaced by 0) and dynamical simulations. The sign composition of organisms differs significantly only in two cases (brown background): in the two versions of loop analysis predictions for site 4 (XLSX 671 kb)
42974_2021_68_MOESM2_ESM.xlsx
The strength of Spearman correlation (ǀρǀ) between some pairs of simulated interaction strength (aij), interaction strength based on food web topology (TIij), node centrality (Ti), sign-agreement based on loop analysis predictions (LApred) and the adjoint value based on loop analysis (LAadj). The only signifiant correlation is set in bold (XLSX 9 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fábián, V. Predicting the sign of trophic effects: individual-based simulation versus loop analysis. COMMUNITY ECOLOGY 22, 441–451 (2021). https://doi.org/10.1007/s42974-021-00068-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42974-021-00068-1