Abstract
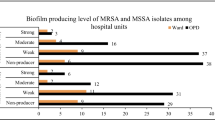
Staphylococcus aureus is a common clinical pathogen that causes various human infections. The aim of this study was to investigate the antibiotic susceptibility pattern, molecular epidemiological characteristics, and biofilm formation ability of S. aureus isolates from clinical specimens in Xiangyang and to analyze the correlation among them. A total of 111 non-duplicate S. aureus isolates were collected from the Affiliated Hospital of Hubei University of Arts and Science. All isolates were tested for antibacterial susceptibility. Methicillin-resistant S. aureus (MRSA) was identified by the mecA gene PCR amplification. All isolates were analyzed to determine their biofilm-forming ability using the microplate method. The biofilm-related gene was determined using PCR. SCCmec, MLST, and spa types of MRSA strains were performed to ascertain the molecular characteristics. Among the 111 S. aureus isolates, 45 (40.5%) and 66 (59.5%) were MRSA and MSSA, respectively. The resistance of MRSA strains to the tested antibiotics was significantly stronger than that of MSSA strains. All isolates were able to produce biofilm with levels ranging from strong (28.9%, 18.2%), moderate (62.2%, 62.1%), to weak (8.9%, 19.7%). Strong biofilm formation was observed in MRSA strains than in MSSA strains, based on percentages. There were dynamic changes in molecular epidemic characteristics of MRSA isolates in Xiangyang. SCCmecIVa-ST22-t309, SCCmecIVa-ST59-t437, and SCCmecIVa-ST5-t2460 were currently the main epidemic clones in this region. SCCmecIVa-ST5-t2460 and SCCmecIVa/III-ST22-t309 have stronger antibiotic resistance than SCCmecIVa-ST59-t437 strains, with resistance to 6 ~ 8 detected non-β-lactam antibiotics. The molecular epidemic and resistance attributes of S. aureus should be timely monitored, and effective measures should be adopted to control the clinical infection and spread of the bacteria.





Similar content being viewed by others
Data Availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Mulvey MR, Simor AE (2009) Antimicrobial resistance in hospitals: how concerned should we be? CMAJ 180(4):408–415. https://doi.org/10.1503/cmaj.080239
Fang H, Fröding I, Gian B, Hæggman S, Tollström UB, Ullberg M, Nord CE (2016) Methicillin-resistant Staphylococcus aureus in Stockholm, Sweden: molecular epidemiology and antimicrobial susceptibilities to ceftaroline, linezolid, mupirocin and vancomycin in 2014. J Glob Antimicrob Resist 5:31–35. https://doi.org/10.1016/j.jgar.2016.01.012
Jin Y, Zhou W, Yin Z, Zhang S, Chen Y, Shen P, Ji J, Chen W, Zheng B, Xiao Y (2021) The genetic feature and virulence determinant of highly virulent community-associated MRSA ST338-SCCmec Vb in China. Emerg Microbes Infect 10(1):1052–1064. https://doi.org/10.1080/22221751.2021.1914516
Bavaro DF, Belati A, Bussini L, Cento V, Diella L, Gatti M, Saracino A, Pea F, Viale P, Bartoletti M (2023) Safety and effectiveness of fifth generation cephalosporins for the treatment of methicillin-resistant staphylococcus aureus bloodstream infections: a narrative review exploring past, present, and future. Expert Opin Drug Saf 25:1–28. https://doi.org/10.1080/14740338.2023.2299377
Madera S, McNeil N, Serpa PH, Kamm J, Pak C, Caughell C, Nichols A, Dynerman D, Li LM, Sanchez-Guerrero E (2023) Prolonged silent carriage, genomic virulence potential and transmission between staff and patients characterize a neonatal intensive care unit (NICU) outbreak of methicillin-resistant Staphylococcus aureus (MRSA). Infect Control Hosp Epidemiol 44(1):40–46. https://doi.org/10.1017/ice.2022.48
Thomsen J, Abdulrazzaq NM; UAE AMR Surveillance Consortium; Menezes GA, Ayoub Moubareck C, Everett DB, Senok A (2023) Methicillin resistant Staphylococcus aureus in the United Arab Emirates: a 12-year retrospective analysis of evolving trends. Front Public Health 11:1244351. https://doi.org/10.3389/fpubh.2023.1244351
Algammal AM, Enany ME, El-Tarabili RM, Ghobashy MOI, Helmy YA (2020) Prevalence, antimicrobial resistance profiles, virulence and enterotoxins-determinant genes of MRSA isolated from subclinical bovine mastitis in Egypt. Pathogens 9(5):362. https://doi.org/10.3390/pathogens9050362
Hou Z, Liu L, Wei J, Xu B (2023) Progress in the prevalence, classification and drug resistance mechanisms of methicillin-resistant Staphylococcus aureus. Infect Drug Resist 16:3271–3292. https://doi.org/10.2147/IDR.S412308
Chen H, Yin Y, van Dorp L, Shaw LP, Gao H, Acman M, Yuan J, Chen F, Sun S, Wang X et al (2021) Drivers of methicillin-resistant Staphylococcus aureus (MRSA) lineage replacement in China. Genome Med 13(1):171. https://doi.org/10.1186/s13073-021-00992-x
Tam K, Torres VJ (2019) Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol Spectr 7(2):10.1128. https://doi.org/10.1128/microbiolspec.GPP3-0039-2018
Azara E, Longheu C, Sanna G, Tola S (2017) Biofilm formation and virulence factor analysis of Staphylococcus aureus isolates collected from ovine mastitis. J Appl Microbiol 123(2):372–379. https://doi.org/10.1111/jam.13502
Achek R, El-Adawy H, Hotzel H, Tomaso H, Ehricht R, Hamdi TM, Azzi O, Monecke S (2020) Short communication: diversity of staphylococci isolated from sheep mastitis in northern Algeria. J Dairy Sci 103(1):890–897. https://doi.org/10.3168/jds.2019-16583
Brady RA, Leid JG, Calhoun JH, Costerton JW, Shirtliff ME (2008) Osteomyelitis and the role of biofilms in chronic infection. FEMS Immunol Med Microbiol 52(1):13–22. https://doi.org/10.1111/j.1574-695X.2007.00357.x
Fitzpatrick F, Humphreys H, O’Gara JP (2005) The genetics of staphylococcal biofilm formation—will a greater understanding of pathogenesis lead to better management of device-related infection? Clin Microbiol Infect 11(12):967–973. https://doi.org/10.1111/j.1469-0691.2005.01274.x
Luther MK, Parente DM, Caffrey AR, Daffinee KE, Lopes VV, Martin ET, LaPlante KL (2018) Clinical and genetic risk factors for biofilm-forming Staphylococcus aureus. Antimicrob Agents Chemother 62(5):e02252-e2317. https://doi.org/10.1128/AAC.02252-17
Yang S, Wang B, Li J, Zhao X, Zhu Y, Sun Q, Liu H, Wen X (2022) Genetic diversity, antibiotic resistance, and virulence gene features of methicillin-resistant Staphylococcus aureus epidemics in Guiyang, Southwest China. Infect Drug Resist 15:7189–7206. https://doi.org/10.2147/IDR.S392434
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 33th ed. Wayne, PA: CLSI; 2023 Document M100
Oliveira DC, de Lencastre H (2002) Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 46(7):2155–2161. https://doi.org/10.1128/AAC.46.7.2155-2161.2002
Stepanović S, Vuković D, Hola V, Di Bonaventura G, Djukić S, Cirković I, Ruzicka F (2007) Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115(8):891–899. https://doi.org/10.1111/j.1600-0463.2007.apm_630.x
Hamad PA (2023) Phenotypic and molecular detection of biofilm formation in methicillin-resistant Staphylococcus aureus isolated from different clinical sources in Erbil City. Mediterr J Hematol Infect Dis 15(1):e2023016. https://doi.org/10.4084/MJHID.2023.016
Tang J, Chen J, Li H, Zeng P, Li J (2013) Characterization of adhesin genes, staphylococcal nuclease, hemolysis, and biofilm formation among Staphylococcus aureus strains isolated from different sources. Foodborne Pathog Dis 10(9):757–763. https://doi.org/10.1089/fpd.2012.1474
Zhang K, McClure JA, Elsayed S, Louie T, Conly JM (2005) Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol 43(10):5026–5033. https://doi.org/10.1128/JCM.43.10.5026-5033.2005
Zhang K, McClure JA, Conly JM (2012) Enhanced multiplex PCR assay for typing of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. Mol Cell Probes 26(5):218–221. https://doi.org/10.1016/j.mcp.2012.04.002
Liang J, Hu Y, Fu M, Li N, Wang F, Yu X, Ji B (2023) Resistance and molecular characteristics of methicillin-resistant Staphylococcus aureus and heterogeneous vancomycin-intermediate Staphylococcus aureus. Infect Drug Resist 16:379–388. https://doi.org/10.2147/IDR.S392908
Zhang H, Cao J, He Z, Zong X, Sun B (2023) Molecular epidemiology of Staphylococcus aureus in a tertiary hospital in Anhui, China: ST59 remains a serious threat. Infect Drug Resist 16:961–976. https://doi.org/10.2147/IDR.S395220
Tang YT, Cao R, Xiao N, Li ZS, Wang R, Zou JM, Pei J (2018) Molecular epidemiology and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolates in Xiangyang, China. J Glob Antimicrob Resist 12:31–36. https://doi.org/10.1016/j.jgar.2017.08.016
Yousefi M, Pourmand MR, Fallah F, Hashemi A, Mashhadi R, Nazari-Alam A (2016) Characterization of Staphylococcus aureus biofilm formation in urinary tract infection. Iran J Public Health 45(4):485–493
Chen Z, Han C, Huang X, Liu Y, Guo D, Ye X (2018) A molecular epidemiological study of methicillin-resistant and methicillin-susceptible Staphylococcus aureus contamination in the airport environment. Infect Drug Resist 11:2363–2375
Rather MA, Gupta K, Bardhan P, Borah M, Sarkar A, Eldiehy KSH, Bhuyan S, Mandal M (2021) Microbial biofilm: a matter of grave concern for human health and food industry. J Basic Microbiol 61(5):380–395. https://doi.org/10.1002/jobm.202000678
Piechota M, Kot B, Frankowska-Maciejewska A, Grużewska A, Woźniak-Kosek A (2018) biofilm formation by methicillin-resistant and methicillin-sensitive Staphylococcus aureus strains from hospitalized patients in Poland. Biomed Res Int 2018:4657396. https://doi.org/10.1155/2018/4657396
Leshem T, Schnall BS, Azrad M, Baum M, Rokney A, Peretz A (2022) Incidence of biofilm formation among MRSA and MSSA clinical isolates from hospitalized patients in Israel. J Appl Microbiol 133(2):922–929. https://doi.org/10.1111/jam.15612
Omidi M, Firoozeh F, Saffari M, Sedaghat H, Zibaei M, Khaledi A (2020) Ability of biofilm production and molecular analysis of spa and ica genes among clinical isolates of methicillin-resistant Staphylococcus aureus. BMC Res Notes 13(1):19. https://doi.org/10.1186/s13104-020-4885-9
Schulte B, Bierbaum G, Pohl K, Goerke C, Wolz C (2013) Diversification of clonal complex 5 methicillin-resistant Staphylococcus aureus strains (Rhine-Hesse clone) within Germany. J Clin Microbiol 51(1):212–216. https://doi.org/10.1128/JCM.01967-12
Miller RM, Price JR, Batty EM, Didelot X, Wyllie D, Golubchik T, Crook DW, Paul J, Peto TE, Wilson DJ et al (2014) Healthcare-associated outbreak of meticillin-resistant Staphylococcus aureus bacteraemia: role of a cryptic variant of an epidemic clone. J Hosp Infect 86(2):83–89. https://doi.org/10.1016/j.jhin.2013.11.007
Ko KS, Lee JY, Suh JY, Oh WS, Peck KR, Lee NY, Song JH (2005) Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J Clin Microbiol 43(1):421–426. https://doi.org/10.1128/JCM.43.1.421-426.2005
Sato T, Yamaguchi T, Aoki K, Kajiwara C, Kimura S, Maeda T, Yoshizawa S, Sasaki M, Murakami H, Hisatsune J et al (2023) Whole-genome sequencing analysis of molecular epidemiology and silent transmissions causing meticillin-resistant Staphylococcus aureus bloodstream infections in a university hospital. J Hosp Infect 139:141–149. https://doi.org/10.1016/j.jhin.2023.05.014
Liu Y, Wang H, Du N, Shen E, Chen H, Niu J, Ye H, Chen M (2009) Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob Agents Chemother 53(2):512–518. https://doi.org/10.1128/AAC.00804-08
Thapaliya D, Forshey BM, Kadariya J, Quick MK, Farina S, O’Brien A, Nair R, Nworie A, Hanson B, Kates A et al (2017) Prevalence and molecular characterization of Staphylococcus aureus in commercially available meat over a one-year period in Iowa, USA. Food Microbiol. 65:122–129. https://doi.org/10.1016/j.fm.2017.01.015
Li X, Zhang J, Zhang Y, Zhou J, Li X, Feng R, Li Y (2021) Methicillin-resistant Staphylococcus aureus of the clonal lineage ST5-SCCmecII-t2460 was associated with high mortality in a Wuhan hospital. Braz J Microbiol 52(4):1929–1936. https://doi.org/10.1007/s42770-021-00557-5
Tan S, Wan C, Wang H, Zhou W, Shu M (2019) Relationship between nasal carrier isolates and clinical isolates in children with Staphylococcus aureus infections. Microb Pathog 127:233–238. https://doi.org/10.1016/j.micpath.2018.11.032
Holden MT, Hsu LY, Kurt K, Weinert LA, Mather AE, Harris SR, Strommenger B, Layer F, Witte W, de Lencastre H et al (2013) A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res 23(4):653–664. https://doi.org/10.1101/gr.147710.112
Zhao H, Wu X, Wang B, Shen L, Rao L, Wang X, Zhang J, Xiao Y, Xu Y, Yu J et al (2023) Phenotypic and genomic analysis of the hypervirulent ST22 methicillin-resistant Staphylococcus aureus in China. mSystems 8(3):e0124222. https://doi.org/10.1128/msystems.01242-22
Ludden C, Brennan G, Morris D, Austin B, O’Connell B, Cormican M (2015) Characterization of methicillin-resistant Staphylococcus aureus from residents and the environment in a long-term care facility. Epidemiol Infect 143(14):2985–2988. https://doi.org/10.1017/S0950268815000072
Chen H, Liu Y, Jiang X, Chen M, Wang H (2010) Rapid change of methicillin-resistant Staphylococcus aureus clones in a Chinese tertiary care hospital over a 15-year period. Antimicrob Agents Chemother 54(5):1842–1847. https://doi.org/10.1128/AAC.01563-09
Asadollahi P, Farahani NN, Mirzaii M, Khoramrooz SS, van Belkum A, Asadollahi K, Dadashi M, Darban-Sarokhalil D (2018) Distribution of the most prevalent Spa types among clinical isolates of methicillin-resistant and -susceptible Staphylococcus aureus around the world: a review. Front Microbiol 9:163. https://doi.org/10.3389/fmicb.2018.00163
Li S, Sun S, Yang C, Chen H, Yin Y, Li H, Zhao C, Wang H (2018) The changing pattern of population structure of Staphylococcus aureus from bacteremia in China from 2013 to 2016: ST239-030-MRSA Replaced by ST59-t437. Front Microbiol 9:332. https://doi.org/10.3389/fmicb.2018.00332
Wang B, Xu Y, Zhao H, Wang X, Rao L, Guo Y, Yi X, Hu L, Chen S, Han L et al (2022) Methicillin-resistant Staphylococcus aureus in China: a multicentre longitudinal study and whole-genome sequencing. Emerg Microbes Infect 11(1):532–542. https://doi.org/10.1080/22221751.2022.2032373
Dai Y, Liu J, Guo W, Meng H, Huang Q, He L, Gao Q, Lv H, Liu Y, Wang Y et al (2019) Decreasing methicillin-resistant Staphylococcus aureus (MRSA) infections is attributable to the disappearance of predominant MRSA ST239 clones, Shanghai, 2008–2017. Emerg Microbes Infect 8(1):471–478. https://doi.org/10.1080/22221751.2019.1595161
Pulingam T, Parumasivam T, Gazzali AM, Sulaiman AM, Chee JY, Lakshmanan M, Chin CF, Sudesh K (2022) Antimicrobial resistance: prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur J Pharm Sci 170:106103. https://doi.org/10.1016/j.ejps.2021.106103
Acknowledgements
We sincerely thank all the participants who took part in this study.
Funding
This study was funded by Xiangyang Science and Technology Research and Development Project [grant no. 2021ZD23].
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
All isolates in this study were collected during bacterial isolation and identification in the clinical microbiology laboratory of the Affiliated Hospital of Hubei University of Arts and Sciences. Patients were treated anonymously, so ethical approval is not required.
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: Nilton Lincopan
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tang, Y., Xiao, N., Zou, J. et al. Antibiotic resistance, biofilm formation, and molecular epidemiology of Staphylococcus aureus in a tertiary hospital in Xiangyang, China. Braz J Microbiol (2024). https://doi.org/10.1007/s42770-024-01270-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42770-024-01270-9