Abstract
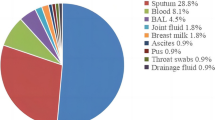
The ica genes in methicillin-resistant Staphylococcus aureus (MRSA) play an important role in biofilm formation. The aim of this study is to define effect of antibiotic resistance and clinical specimens to the expression of ica genes based on their sequence types (STs) and clonal complex (CC). One-hundred (100) S. aureus strain were collected from two teaching therapeutic centers in Hamedan, Iran. Then, the PCR, qPCR, and MLST were used to characterize strains. The results indicated that 29 (29%), 15 (15%), and 5 (5%) strain were strong, mediate, weak biofilm producer, respectively, and the icaA (17%) and icaC (14%) genes were the most abundant. However, two unique STs (3667, 491) in Iran were reported and ST30 and ST11 were the most abundant STs and CC30 and CC5 were observed among MRSA and MSSA strains. High activity in ica locus was observed among strains collected from wound and catheter strains. Also, expression level of icaA gene increased in all strains except ST30 and ST491. Moreover, the highest expression level was observed in CC1, CC7, and CC11. Likewise, activity of the icaC gene was only observed in CC5. Furthermore, the expression of all ica genes in CC5 was significantly correlated with the type of biofilm and the clinical sample. In this study demonstrated that the frequency distribution of STs and CCs in different strains of MRSA was higher than methicillin-sensitive strains. Also, the type of clinical specimen and expression of ica genes played an important role in this abundance.




Similar content being viewed by others
References
Heydari N, Alikhani MY, Jalilian FA et al (2017) Evaluation of real time PCR for detection of clinical isolates of Staphylococcus aureus and methicillin-resistance strains based on melting curve analysis method. Koomesh 19(4):877–886
Pasandideh NK, Habibi MR, Tahmasebi H et al (2018) Activity of biofilm genes icaA and icaR in methicillin-resistant Staphylococcus aureus treated with vitamin K in wound specimens. Koomesh 20(3):588–593
Boswihi SS, Udo EE, Al-Sweih N (2016) Shifts in the clonal distribution of methicillin-resistant Staphylococcus aureus in Kuwait hospitals: 1992–2010. PLoS ONE 11(9):e0162744
Udo EE, Boswihi SS (2017) Antibiotic resistance trends in methicillin-resistant Staphylococcus aureus Isolated in Kuwait Hospitals: 2011–2015. Med Princ Pract 26(5):485–490
Vafaeefar M, Yousef Alikhani M, Tahmasebi H et al (2017) Identification and determination of the relationship between ccr alleles and antibiotic resistance in clinical isolates of methicillin resistant Staphylococcus aureus. J Babol Univ Med Sci 19(12):28–35
Bongiorno D, Mongelli G, Stefani S et al (2017) Burden of rifampicin- and methicillin-resistant Staphylococcus aureus in Italy. Microbial Drug Resist 24(6):732–738
Sritharadol R, Hamada M, Kimura S et al. (2018). Mupirocin at subinhibitory concentrations induces biofilm formation in Staphylococcus aureus. Microb Drug Resist.
Ghasemian A, Najar-Peerayeh S, Bakhshi B et al (2015) High prevalence of icaABCD genes responsible for biofilm formation in clinical isolates of Staphylococcus aureus from hospitalized children. Arch Pediatr Infect Dis 3(3):e20703
McCarthy H, Rudkin JK, Black NS et al (2015) Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front Cell Infect Microbiol 5:1–1
Dehbashi S, Tahmasebi H, Zeyni B et al (2018) The relationship between promoter-dependent quorum sensing induced genes and methicillin resistance in clinical strains of Staphylococcus aureus. J Zanjan Univ Med Sci 26(116):75–87
da Fonseca W, Batistão D, Amaral de Campos P, Caroline Camilo N et al (2016) Biofilm formation of Brazilian meticillin-resistant Staphylococcus aureus strains: prevalence of biofilm determinants and clonal profiles. J Med. Microbiol 65(4):286–297
Long SW, Olsen RJ, Mehta SC et al (2014) PBP2a Mutations causing high-level ceftaroline resistance in clinical methicillin-resistant Staphylococcus aureus isolates. Antimicrob Agents Chemother 58(11):6668–6674
Teixeira MM, Araújo MC, Silva-Carvalho MC et al (2012) Emergence of clonal complex 5 (CC5) methicillin-resistant Staphylococcus aureus (MRSA) isolates susceptible to trimethoprim-sulfamethoxazole in a Brazilian hospital. Br J Med Biol Res 45(7):637–643
setia m s. (2016) Methodology series module 5: sampling strategies. Indian J Pediatr 61(5):505–509
Bokaeian M, Tahmasebi H (2017) Molecular identification of genes responsible for resistance to aminoglycosides and methicillin in clinical samples of Staphylococcus aureus. J Babol Univ Med Sci 19(3):38–46. https://doi.org/10.22088/jbums.19.3.38
Clinical and Laboratory Standards Institute (2017) Performance standards for antimicrobial susceptibility testing. twenty-seventh informational supplement. M100S, 27 edn. CLSI, Wayne
Côrtes MF, Beltrame CO, Ramundo MS et al (2015) The influence of different factors including fnbA and mecA expression on biofilm formed by MRSA clinical isolates with different genetic backgrounds. Int J Med Microbiol 305(1):140–147
Solati SM, Tajbakhsh E, Khamesipour F et al (2015) Prevalence of virulence genes of biofilm producing strains of Staphylococcus epidermidis isolated from clinical samples in Iran. AMB Express 5(1):47
Strommenger B, Kettlitz C, Weniger T et al (2006) Assignment of staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J Clin Microbiol 44(7):2533–2540
Abubakar U, Sulaiman SAS (2018) Prevalence, trend and antimicrobial susceptibility of methicillin resistant Staphylococcus aureus in Nigeria: a systematic review. Int J Infect Control 11(6):763–770
Khosravi AD, Jenabi A, Montazeri EA (2017) Distribution of genes encoding resistance to aminoglycoside modifying enzymes in methicillin-resistant Staphylococcus aureus (MRSA) strains. Kaohsiung J Med Sci 33(12):587–593
Sit PS, Teh CSJ, Idris N et al (2017) Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) infection and the molecular characteristics of MRSA bacteraemia over a two-year period in a tertiary teaching hospital in Malaysia. BMC Infect Dis 17(1):274–274
Ravensbergen S J, Berends M, Stienstra Y et al. (2017). High prevalence of MRSA and ESBL among asylum seekers in the Netherlands. PLoS ONE, 12(4), e0176481.
Rağbetli C, Parlak M, Bayram Y et al (2016) Evaluation of antimicrobial resistance in Staphylococcus aureus isolates by years. Jpn J Infect Dis 2016:9171395–9171395
Horner C, Wilcox M, Barr B et al. (2012). The longitudinal prevalence of MRSA in care home residents and the effectiveness of improving infection prevention knowledge and practice on colonisation using a stepped wedge study design. BMJ Open, 2(1).
Loughman JA, Fritz SA, Storch GA et al (2009) Virulence gene expression in human community-acquired Staphylococcus aureus infection. Int J Infect Dis 199(3):294–301
Algburi A, Comito N, Kashtanov D et al. (2017). Control of biofilm formation: antibiotics and beyond. Appl Environ Microbiol, 83(3).
Jimi S, Miyazaki M, Takata T et al (2017) Increased drug resistance of meticillin-resistant Staphylococcus aureus biofilms formed on a mouse dermal chip model. J Med Microbiol 66(4):542–550
Cerca N, Brooks JL, Jefferson KK (2008) Regulation of the intercellular adhesin locus regulator (icaR) by SarA, sigmaB, and IcaR in Staphylococcus aureus. J bacteriol 190(19):6530–6533
Serray B, Oufrid S, Hannaoui I et al (2016) Genes encoding adhesion factors and biofilm formation in methicillin-resistant Staphylococcus aureus in Morocco. J Infect Dev Ctries 10(8):863–869
Kot B, Sytykiewicz H, and Sprawka I. (2018). Expression of the Biofilm-Associated Genes in Methicillin-Resistant Staphylococcus aureus in Biofilm and Planktonic Conditions. Int J Mol Sci, 19(11).
Piechota M, Kot B, Maciejewska AF et al (2018) Biofilm formation by methicillin-resistant and methicillin-sensitive Staphylococcus aureus strains from hospitalized patients in Poland. Biomed Res Int 2018:7
Tasse J, Trouillet-Assant S, Josse J et al (2018) Association between biofilm formation phenotype and clonal lineage in Staphylococcus aureus strains from bone and joint infections. PLoS ONE 13(8):e0200064–e0200064
Vanhommerig E, Moons P, Pirici D et al (2014) Comparison of biofilm formation between major clonal lineages of methicillin resistant Staphylococcus aureus. PLoS ONE 9(8):e104561–e104561
Challagundla L, Reyes J, Rafiqullah I et al (2018) Phylogenomic classification and the evolution of clonal complex 5 methicillin-resistant Staphylococcus aureus in the western hemisphere. Front Microbiol 9:1901–1901
Marbach H, Boakes E, Lynham S et al (2017) Identification of a distinctive phenotype for endocarditis-associated clonal complex 22 MRSA isolates with reduced vancomycin susceptibility. J Med Microbiol 66(5):584–591
Acknowledgements
The authors of this article are grateful to Hamadan University of Medical Sciences for their financial support in conducting research.
Author information
Authors and Affiliations
Contributions
MRZ designed research; HT, SD and MJ performed experiments. HT and MRA wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tahmasebi, H., Dehbashi, S., Jahantigh, M. et al. Relationship between biofilm gene expression with antimicrobial resistance pattern and clinical specimen type based on sequence types (STs) of methicillin-resistant S. aureus. Mol Biol Rep 47, 1309–1320 (2020). https://doi.org/10.1007/s11033-019-05233-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-019-05233-4