Abstract
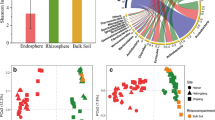
To study the correlation between root microbiome and its community structure and the health, survival, and growth of its host which is the key to solve the problem of the diversity relationship between wild plants and root microorganism. In this study, we used high-throughput techniques of next generation sequencing (NGS) which was applied to study the endophytic and rhizosphere bacterial and fungal community in hulless barley (Hordeum vulgare) plants, by assessing its PCR amplicon of 16S rDNA sequences and ITS region. The results of the principal component analysis (PCA) showed that bacterial phyla Proteobacteria, Actinobacteria, and Acidobacteria dominate the bacterial community and that the phyla of Ascomycota and Basidiomycota dominate the mycobiota community in the root-soil interface of hulless barley. In both 16S and ITS data, the alpha diversity in bulk soil samples was significantly higher than that of rhizosphere and root samples, and root sample was least diverse, suggesting the microbial selection from the plant host. Beta diversity analysis indicated a clear separation from samples with different sample types (bulk soil, rhizosphere, and root samples). Lastly, the overall microbiota profile and differentially presented taxa were studied to assess the function. It can be concluded that the microbial diversity of wild hulless barley in different soil samples was significantly different and related to host genotypes.




Similar content being viewed by others
References
Achatz B, von Rüden S, Andrade D, Neumann E, Pons-Kühnemann J, Kogel K-H, Franken P, Waller F (2010) Root colonization by Piriformospora Indica enhances grain yield in barley under diverse nutrient regimes by accelerating plant development. Plant Soil 333(1–2):59–70
Anderson MJ, Walsh DCI (2013) PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol Monogr 83(4):557–574
Anzai Y, Kim H, Park JY, Wakabayashi H, Oyaizu H (2000) Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol 50(Pt 4):1563–1589
Bahulikar RA, Torres-Jerez I, Worley E, Craven K, Udvardi MK (2014) Diversity of nitrogen-fixing bacteria associated with switchgrass in the native tallgrass prairie of Northern Oklahoma, ed. J. E. Kostka. Appl Environ Microbiol 80(18):5636–5643
Bhatty RS (1999a) The potential of hull-less barley. Cereal Chem 76(5):589–599
Bhatty RS (1999b) β-Glucan and flour yield of hull-less barley. Cereal Chem 76(2):314–315
Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, McHardy AC, Schulze-Lefert P (2015) Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 17(3):392–403
Caporaso JG et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Chen X, Long H, Gao P, Deng G, Pan Z, Liang J, Tang Y, Tashi N, Yu M (2014) Transcriptome assembly and analysis of Tibetan hulless barley (Hordeum Vulgare L. Var. Nudum) developing grains, with emphasis on quality properties. PLoS One 9(5):e98144
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72(7):5069–5072
Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J (2014) An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2(1):6
Gottel NR, Castro HF, Kerley M, Yang Z, Pelletier DA, Podar M, Karpinets T, Uberbacher E, Tuskan GA, Vilgalys R, Doktycz MJ, Schadt CW (2011) Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl Environ Microbiol 77(17):5934–5944
Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, Döring M, Sessitsch A (2015) The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev 79(3):293–320
Hauben L, Vauterin L, Moore ERB, Hoste B, Swings J (1999) Genomic diversity of the genus Stenotrophomonas. Int J Syst Bacteriol 49(Pt 4):1749–1760
Jones DL, Hodge A, Kuzyakov Y (2004) Plant and mycorrhizal regulation of rhizodeposition. New Phytol 163(3):459–480
Kaul S, Sharma T, K. Dhar M (2016) ‘Omics’ tools for better understanding the plant–endophyte interactions. Front Plant Sci 7
Kogel K-H, Franken P, Hückelhoven R (2006) Endophyte or parasite--what decides? Curr Opin Plant Biol 9(4):358–363
Kõljalg U, Larsson K-H, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E, Pennanen T, Sen R, Taylor AFS, Tedersoo L, Vrålstad T (2005) UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytol 166(3):1063–1068
Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31(9):814–821
Latch GCM (1993) Physiological interactions of endophytic fungi and their hosts. Biotic stress tolerance imparted to grasses by endophytes. Agric Ecosyst Environ 44(1–4):143–156
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556
Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, Rio TG d, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL (2012) Defining the core Arabidopsis Thaliana root microbiome. Nature 488(7409):86–90
Mandal S et al (2015) Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb Ecol Health Dis 26:27663
McMurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data, ed. Michael Watson. PLoS One 8(4):e61217
Miliute I, Buzaite O, Baniulis D, Stanys V (2015) Bacterial endophytes in agricultural crops and their role in stress tolerance: a review. Zemdirbyste-Agriculture 102(4):465–478
Murphy BR, Doohan FM, Hodkinson TR (2014) Fungal endophytes of barley roots. J Agric Sci 152(4):602–615
Nagashima S, Kamimura A, Shimizu T, Nakamura-Isaki S, Aono E, Sakamoto K, Ichikawa N, Nakazawa H, Sekine M, Yamazaki S, Fujita N, Shimada K, Hanada S, Nagashima KVP (2012) Complete genome sequence of phototrophic betaproteobacterium Rubrivivax Gelatinosus IL144. J Bacteriol 194(13):3541–3542
Narwal S, Kumar D, Sheoran S, Verma RPS, Gupta RK (2017) Hulless Barley as a promising source to improve the nutritional quality of wheat products. J Food Sci Technol 54(9):2638–2644
Schreiber M et al (2014) The barley genome sequence assembly reveals three additional members of the CslF (1,3;1,4)-β-glucan synthase gene family, ed. Samuel P. Hazen. PLoS One 9(3):e90888
Sengupta S, Ganguli S, Singh PK (2017) Metagenome analysis of the root endophytic microbial community of Indian rice ( O. Sativa L.). Genomics Data 12:41–43
Tkacz A, Poole P (2015) Role of root microbiota in plant productivity. J Exp Bot 66(8):2167–2175
van der Heijden MGA, Bardgett RD, van Straalen NM (2008) The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol Lett 11(3):296–310
Yang P et al (2008) Genetic diversity analysis of the developed qingke (hulless barley) cultivars from the plateau regions of Sichuan province in China revealed by SRAP markers. Yi Chuan 30(1):115–122
Zeng XQ (2015) Genetic variability in agronomic traits of a germplasm collection of hulless barley. Genet Mol Res 14(4):18356–18369
Zeng X, Long H, Wang Z, Zhao S, Tang Y, Huang Z, Wang Y, Xu Q, Mao L, Deng G, Yao X, Li X, Bai L, Yuan H, Pan Z, Liu R, Chen X, WangMu QM, Chen M, Yu L, Liang J, DunZhu DW, Zheng Y, Yu S, LuoBu ZX, Guang X, Li J, Deng C, Hu W, Chen C, TaBa XN, Gao L, Lv X, Abu YB, Fang X, Nevo E, Yu M, Wang J, Tashi N (2015) The draft genome of Tibetan hulless barley reveals adaptive patterns to the high stressful Tibetan Plateau. Proc Natl Acad Sci U S A 112(4):1095–1100
Funding
This study is funded by the program of Junior Teacher Innovation Project of Tibet Autonomous Region (QCZ2016-45) and the Natural Science Foundation of Tibet Autonomous Region (2015212-14-23).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Figure 1
(TIFF 1.11 mb)
Supplementary Table 1
(CSV 4.11 kb)
Supplementary Table 2
(CSV 1.54 mb)
Rights and permissions
About this article
Cite this article
Liu, Z.D., Li, L., Zhuo, G. et al. Characterizing Structure and Potential Function of Bacterial and Fungal Root Microbiota in Hulless Barley Cultivars. J Soil Sci Plant Nutr 19, 420–429 (2019). https://doi.org/10.1007/s42729-019-00045-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42729-019-00045-8