Abstract
Dengue virus, a pervasive mosquito-borne pathogen, imposes a substantial global health burden and is responsible for numerous fatalities annually globally, with tropical and sub-tropical regions particularly susceptible to dengue outbreaks. Despite decades of efforts, there has been no effective treatment or prevention for dengue, which makes it a life-threatening disease. Hence, this study proposes an innovative bioinformatics-driven approach to construct a vaccine targeting the dengue virus. The study involved a comprehensive analysis of conserved regions of dengue virus serotypes 1–4's non-structural proteins (NS1, NS3, and NS5) and structural protein (E) to predict the potential B & T-cell epitopes which were linked with appropriate adjuvants and linkers to generate four distinct vaccine candidates. The constructed vaccine models underwent rigorous evaluation, considering physicochemical attributes, structural integrity, population coverage, and immune system response through simulation. The results confirm that these vaccine candidates are non-allergenic, non-toxic, antigenic, and immunogenic. Additionally, they exhibit 99.70% world population coverage and 100% conservation across all dengue strains, which is crucial for vaccine efficacy. A Ramachandran plot showed that 95.6% of the amino acid residues of the candidates belong to the optimal zone, while around 4% are in additional allowed regions. Further, molecular docking and dynamic simulation of interaction with the human toll-like receptor 4, a fundamental component of innate immunity, was carried out to gain more insight into interaction dynamics. As a result of these analyses, the candidates' binding dynamics and structural stability were revealed. Overall, this study presents promising vaccine candidates for addressing dengue's global health burden. Their robust design and demonstrated immunogenicity make them attractive candidates for further experimental testing and development as potential vaccines against current strains and future variants.
Article highlights
-
1.
Innovative bioinformatics-driven strategy leads to multi-epitope dengue vaccine development.
-
2.
The vaccine candidates are predicted to be safe, effective against all strains, and cover 99.70% of the global population.
-
3.
Molecular simulations provide key insights into vaccine's structural integrity and receptor interactions.
Similar content being viewed by others
1 Introduction
The Dengue virus is a member of the Flaviviridae family and causes severe dengue disease worldwide. There are over a hundred countries where the disease is prevalent [1]. It is most prevalent in the tropical and subtropical regions of America, Africa, Southeast Asia, and the Pacific Ocean [2]. Dengue fever infections claim the lives of nearly 36,000 people every year and affect about 390 million people globally. Some countries have witnessed a significant number of cases, such as Bangladesh with 101,000 cases, Vietnam with 320,000 cases, Malaysia with 131,000 cases, and the Philippines with 420,000 cases [3]. The virus can cause two major and sometimes fatal complications: dengue hemorrhagic fever and dengue shock syndrome, which involve DENV-1, DENV-2, DENV-3, and DENV-4 serotypes. Other symptoms include severe headache, stomachache, rash, fatigue, loss of appetite, diarrhea, vomiting, and a bad taste. The virus can also impair the physical and mental development of the infected individuals [2].
Dengue virus (DENV) is characterized by its positive-strand RNA composition. The virus’s 11 kilobase RNA genome encodes three structural proteins: the capsid, pre-membrane, and envelope, and seven non-structural proteins: NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. These proteins are initially translated as a single polyprotein and then cleaved by viral and host cellular proteases at specific sites. One of the non-structural proteins is NS protein 5 or NS5, which plays a role in protecting the viral genome and enhancing polyprotein translation. NS5 is involved in two functions: RNA methyltransferase (MTase) and viral RNA replication. In humans, NS5 interferes with the host’s antiviral interferon signalling by interacting with proteins in the JAK-STAT pathway, among others [4,5,6].
Till date, there is no antiviral drug that can effectively treat dengue virus infections, which cause dengue fever and its related symptoms, such as pain, muscle aches, and fever. Therefore, acetaminophen and paracetamol are commonly used to relieve these symptoms [2]. (CDC, 2021). Researchers attempted to develop a dengue vaccine in 1929 using immunogenic, inactivated viral components such as phenol, formalin, or bile, but they failed to achieve their goal [2]. Later, during WWII, several research facilities produced live-attenuated virus (LAV) vaccines against both DENV type 1 and 2. However, these early practices were stopped due to safety issues, as they involved introducing live viruses into humans [7,8,9]. There have been several new dengue vaccine concepts evaluated in humans and animal models since the 1970s, but none have reached clinical or preclinical success [10], except for the CYD-TDV developed by Sanofi Pasteur. In the US, this vaccine is the only one approved by the CDC and FDA and is available for purchase as Dengvaxia. Clinical trials are also ongoing for Vaxfectin [11], DENVax [12], and TV003 [12]. A number of limitations, however, make it difficult to achieve broad coverage and elicit a robust immune response against dengue virus serotypes [11,12,13,14]. Moreover, development of vaccine from scratch completely following traditional methods is always challenging, time consuming, and costly which rarely succeed [15]. Considering the limitations of the former approach to predict and design dengue vaccine, immunoinformatics emerges as a pivotal player, revolutionizing the landscape of vaccine prediction and design. Immunoinformatics proves instrumental in dissecting the complexity, offering computational tools to analyze genetic diversity, predict epitopes on viral proteins, and assess potential cross-reactivity with diverse viral strain [16]. This approach not only expedites the identification of antigens for vaccine candidates but also ensures the development of vaccines with broad-spectrum protection. Therefore, researchers have capitalized on these advantages, designing numerous dengue vaccines targeting different protein [17].
The development of a functional dengue vaccine has been challenged by antibody-dependent enhancement, an important obstacle to vaccine development. When antibodies fail to neutralize or weakly neutralize the virus, it can infect host cells, leading to increased viral replication and aggravated disease. Cross-reactive antibodies generated by previous exposure to different DENV serotypes or related flaviviruses, such as Zika and yellow fever, can trigger this complex phenomenon [18, 19]. Therefore, we need a vaccine that can protect against each variant of the dengue virus and sustain that protection for an extended period of time to fight the disease and lower the risk of ADE [20, 21]. Multi-epitope vaccines represent a promising advancement in vaccination technology, offering numerous advantages over conventional methods. These vaccines utilize short antigenic peptide fragments, or epitopes, to stimulate specific immune responses while minimizing the risk of allergic reactions [22]. By incorporating multiple epitopes, multi-epitope vaccines can effectively target a broad range of pathogens, including viruses and tumors. One of the key advantages of multi-epitope vaccines is their ability to induce comprehensive immune responses. These vaccines activate cytotoxic T lymphocytes (CTL), helper T cells (Th), and B cells simultaneously, leading to robust cellular and humoral immunity. This multifaceted approach enhances the vaccine's effectiveness in combating infectious diseases and cancers [23]. Furthermore, multi-epitope vaccines offer resistance to genetic variation, ensuring efficacy against evolving pathogens. Their safety and stability profile are superior to conventional vaccines, reducing the risk of adverse effects. Additionally, the streamlined manufacturing and distribution processes make multi-epitope vaccines more accessible and cost-effective [23][23]. There have been many successful applications of computational techniques for developing multi-epitope vaccines for challenging diseases, such as SARS-CoV-2 [25, 26], H. pylori [27]. For these vaccines, it is crucial to choose epitopes with desirable properties, such as immunogenicity, antigenicity, non-allergenicity, safety, and stability across strains.
This study presents a bioinformatics-based dengue vaccine candidate with multiple epitopes. We analyzed structural protein E and nonstructured proteins NS1, NS3, and NS5 across all DENV serotypes to identify potential T and B cell epitopes. We then carefully combined these epitopes with appropriate linkers and adjuvants to form four distinct vaccine candidates. Our findings included physicochemical properties, structural stability, population coverage, and immune response evaluations. In addition, we examined the complex interaction between the vaccine candidates and innate immunity using molecular docking and dynamic simulations. Therefore, our research suggests that a bioinformatics-based vaccine development strategy can provide a strong foundation for developing vaccines. Using this method, we can effectively address the complex genetic evolution and structural variations of the virus, which may prove helpful in addressing an ever-evolving global health threat.
2 Methodology
2.1 Data retrieval of dengue virus polypeptide sequences and phylogenetic tree construction
Several previous studies provided the basis for selecting vaccine targets [16, 28,29,30]. Out of four, three NS proteins (NS1, NS3, and NS5) were selected for t-cell epitopes and one structural protein (E protein) was selected to predict b-cell epitopes. In order to collect a large dataset of polypeptide sequences from around the world, NCBI and ViPR databases were used and filtered for completeness and redundancy. A key step in developing a comprehensive multi-epitope vaccination method was to find conserved peptides within the polyproteins of all dengue serotypes therefore all the filtered sequences were aligned with the reference NS1, NS3, and NS5 proteins of Dengue virus from RefSeq server using the MEGA11 [31] and the Muscle algorithm. Finally, we used the neighbor-joining method in the MEGA11 to construct the phylogenetic tree which was visualized and annotated using the iTOL (Fig. 1).
Overview of the study methodology of in silico vaccine design against dengue virus (type1-4). The iterative process encompasses diverse computational steps, including data retrieval (NCBI, ViPR), phylogenetic analysis, sequence alignment, epitope prediction, screening, vaccine construction, characteristics assessment, molecular docking, molecular simulation, and immune simulation
2.2 Common epitope identification and validation
Potential T- and B-cell epitopes were predicted using the IEDB tool [32] at https://www.iedb.org/home_v3 after sequence alignment. The recommended NetMHCPan 4.1 EL method was used with default epitope lengths for CD8 + and CD4 + epitopes. B-cell epitope length was between 8 and 16 amino acids, and methods such as Bepipred, Bepipred 2.0, and Emini surface accessibility were employed. Finally, we used the ElliPro server to find discontinuous B-cell epitopes [33]. Epitopes common to all forty strains were extracted for each protein type. The IEDB server was used to perform conservancy analysis on predicted MHC class I and II epitopes.
2.3 Allergenicity, antigenicity profiling & physiochemical properties evaluation
The physicochemical characteristics of both the viral proteins and the designed vaccine candidates were assessed using the ProtParam web server [34]. Toxicity and antigenicity of predicted peptides and the final vaccine candidates were assessed with ToxinPred [35] and Vaxijen v2.0 [36]. Allergenicity and IFNγ induction potential of the epitopes were evaluated using AllerTOP v.2.0 [37], and IFNepitope. TNFepitope (http://crdd.osdd.net/raghava/ifnepitope/predict.php) and IL4Pred (https://webs.iiitd.edu.in/raghava/il4pred/) were used to test the ability of MHC-I-related and MHC-II-related epitopes to produce TNF-α and IL-4, respectively.
2.4 Screening of epitopes for autoimmunity and immunogenicity
To design multiepitope vaccines that enhance immunity without causing autoimmunity, it is crucial to select epitopes that are immunogenic and have minimal similarity to human proteins. The NCBI BlastP (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to evaluate the selected epitopes for the possibility of autoimmunity. Moreover, their immunogenicity was assessed using a Class I Immunogenicity prediction tool (http://tools.iedb.org/immunogenicity/).
2.5 Vaccine construction & population coverage analysis
Using amino acid linkers such as AAY, GPGPG, and KK, the selected CTL, HTL, and B-cell epitopes were combined to make the chimeric vaccine. Several vaccines were produced, each with a different adjuvant, such as 50S ribosomal protein L7/L12 and HBD3 with the RR motif + RS09 using the EAAAK linker. In order to boost the vaccine's effectiveness and strength, a synthetic pan DR (PADRE) epitope (AKVAAWTLKAAAC) that activates CD4 + T-cells was added. The PADRE sequence is much more potent than the common T helper epitopes and can overcome the variation in HLA-DR molecules in different individuals. Compared to the vaccines without the PADRE, all vaccines containing the PADRE triggered stronger CTL responses. Additionally, all adjuvant proteins were found to stimulate multiple TLR complexes, resulting in a strong immune activation and CTL orientation. Furthermore, We employed the Immune Epitope Database (IEDB) tools (http://tools.iedb.org/population/) to examine the distribution of helper T lymphocyte (HTL) and cytotoxic T lymphocyte (CTL) epitopes among different populations. By devising these highly customised algorithms specifically for epitope-based vaccination, we were able to minimise complexity and variability across various ethnic populations while maximising research coverage by using IEDB reference alleles. We used these techniques to evaluate potential population coverage and comprehend the distribution of CTL and HTL epitopes in various populations. Using this method allowed us to calculate the percentage of people on the globe with HLA alleles that are able to identify and react to particular epitopes that are part of our vaccination.
2.6 Vaccine 2D and 3D structure prediction, refinement, and validation
The secondary structure of the vaccine formulation was predicted using the freely available PSIPRED server. To obtain insights into the 3D structure, the publicly accessible 3D pro web service [38] was employed. This tool not only predicts the 3D conformation but also sheds light on solvent accessibility, disordered regions, secondary structures, binding sites, and overexpressed solubility. ChimeraX software [39] facilitated the initial 3D structure assessment, followed by refinement via the Galaxy Refine server [40]. Notably, Galaxy Refine significantly enhanced the model's structure and overall quality. Validation of the improved 3D structure was performed using two open-source online tools, PROSA-web [41] for summarizing error scores and PROCHECK (UCLA-DOE LAB) for analyzing the Ramachandran plot.
2.7 Molecular docking analysis
We aimed to target human toll-like receptor 4 (hTLR-4), which has the PDB ID 4G8A, with our multi-epitope vaccine. Using its default settings, we used the ClusPro 2.0 server to analyze the docking interaction between vaccine candidates and receptors [42]. This server predicts optimal docking models by eliminating steric clashes, clustering the 1000 lowest energy structures, and generating numerous conformations.
2.8 Molecular dynamic (MD) simulation
The protein–ligand complexes were simulated for 100 ns using GROMACS version 2020.6 [43]. The CHARMM36m force field and TIP3 water model were used to create a water box with 1 nm edges from the protein surface. By adding ions, the system was neutralized. The system was equilibrated in NVT and NPT ensembles before energy minimization was achieved. With periodic boundary conditions and 2 fs time steps, a 100 ns MD simulation was carried out. The trajectory data was examined every 100 ps. After the simulation, GROMACS' built-in modules (RMSD, RMSF, Gyrate, SASA, and Hbond) were used to do RMSD, RMSF, Rg, and SASA analyses. The analyses were visualized using ggplot2 in RStudio. The MD simulations were done on high-performance Ubuntu 20.04.4 LTS systems at the Bioinformatics Division of the National Institute of Biotechnology.
2.9 Immune simulation
We employed the C-ImmSim agent-based modeling service [44] to simulate the immune responses elicited by our vaccine. C-ImmSim integrates machine learning techniques and position-specific scoring matrices (PSSM) for accurate immune epitope prediction, enabling comprehensive exploration of potential interactions between the vaccine and the immune system. To ensure consistency and comparability with established protocols, we maintained all default simulation parameters. We administered three vaccine injections at 14-week intervals to mirror real-world vaccination practices and evaluate long-term immunological responses. Each dose contained 1000 vaccine particles in both situations, and the simulation was run for 1095-time steps (350 days). Within the simulated timeframe (1, 84, and 168), each time step represents eight hours, with the initial injection taking place at time zero. This time scale effectively captures the dynamic nature of immune responses while maintaining computational efficiency. Initializing the simulation at time zero establishes baseline conditions for assessing subsequent immune reactions [44].
2.10 Codon optimization and In silico cloning
Since the codons used by the E. Coli host system differ from those used by humans, codon optimization is required to boost expression in the bacterial host system. JAVA Codon Adaptation Tool (http://www.jcat.de/) was utilized to optimize the code (Grote et al. 2005), and the E. Coli k12 strain was chosen as the bacterial host system [16]. Additional option of ‘avoid the rho-independent transcription termination’, ‘avoid prokaryote ribosome binding site’, and ‘avoid restriction enzymes cleavage sites’ were all checked. JCAT output includes code that has been optimized in addition to percentages of GC and CAI, which are used to assess the protein expression. Ideal range for CAI value is between 0.8–1.00 and for GC content percentage is between 30 and 70% [45]. XhoI and BstAPI restriction sites were added to N terminal and C terminal respectively to DV-1 and DV-3 vaccine whereas for DV-2 and DV-4 MluI was added to the C terminal instead of BstAPI. Subsequently, the SnapGene tool was used to individually insert each fragment into the pET-28a ( +) vector which was collected from the ‘Addgene’ vector database [46].
3 Result
3.1 Retrieval of polypeptide sequences and phylogenetic analysis
After extensive screening forty polypeptide sequences were selected from distinct parts of the world for each of the four dengue serotypes, as shown in Supplementary Table 1. We used ten sequences to represent each Dengue virus serotype and made a phylogenetic tree using the full sequences of all forty polyproteins, as seen in Fig. 2 which disclosed the complex genetic relationships among the different Dengue virus types and some cases where genetic similarity was not affected by geographical distance. For example, Dengue Virus Type 1 strains from the Pacific and Asia were genetically similar. We also found a separate genetic cluster that included strains from Brazil, Italy, Singapore, and America, which were genetically related even though they were from different world regions. The phylogenetic tree also revealed the unusual and interesting presence of Dengue Virus serotype 2 and 3 strains in the Dengue Virus Type 1 clade, which helps us to better understand the complicated genetic evolution of this virus.
We got the reference sequences of the structural envelope protein and the NS proteins (NS1, NS3, and NS5) and aligned them with the 40 polypeptide sequences. We used these alignments and the consensus-like sequences of all the proteins for the T-cell and B-cell epitope prediction. Figure 3 shows a schematic diagram of the dengue virus polyprotein.
Analysis of the Polyprotein Landscape in Dengue Virus. DENV Genome Reveals 3 Structural Proteins (Envelope, Pre-membrane, and Capsid) and 7 NS Proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5), initially translated as a single polyprotein. The Consensus Sequence was Derived from 40 Distinct Sequences of Various Strains Deposited on NCBI and ViPR
3.2 Selection of CD4 + and CD8 + T Cell Epitopes
We analyzed all the protein sequences and found more than 48,600 different epitopes. Only five MHC-1 epitopes from NS1 were consistent across all sequences, but they were similar to human proteins and discarded. We also found twenty common epitopes for NS3, but only ten were antigenic while only five of them—TVWFVPSIK, KTVWFVPSIK, KTVWFVPSI, GLYGNGVVT, and GKTVWFVPSI—met the criteria of being safe, immunogenic, different from human proteins, and fully conserved. These epitopes also had extensive population coverage, which indicates their ability to activate the immune system. Similarly, we found ten shared MHC-1 epitopes for NS5. Only two of them—DVVPMVTQMA and FCSHHFHEL—were safe, non-toxic, and not similar to human proteins. These epitopes were fully conserved across all NS5 protein sequences, which shows their importance for future dengue treatments (Table 1).
We also examined helper T-cell epitopes (HTLs) using the same protein sequences from all serotypes and found almost 23,600 unique epitopes. Only three of them were common among all the predicted NS1 protein epitopes (Table 2). However, we had to exclude these three frequent epitopes because they were allergic. For the NS3 protein, there were no common epitopes among the strains from all four serotypes. However, we found three epitopes shared by three dengue serotypes 1, 2 and 3. (Table 2). The only non-allergenic and non-toxic was the EAKMLLDNINTPEGI epitope, which is rare in dengue virus type 4 strains. It had the potential to produce cytokines like INF-γ and IL-4. It also showed 100% conservation across all the sequences from dengue serotypes 1, 2 and 3. We included epitopes from serotype 4 strains to induce a balanced immunological response. We found 39 shared epitopes from the 10 different strain sequences we used. Only ten of them were non-allergenic. Surprisingly, none of these epitopes were similar to human protein, and they all activated IFN-γ. We also found 15 common epitopes among all the 40 sequences of the NS5 protein. Eight of them were antigenic. These eight epitopes were not similar to human proteins, safe, and not allergic.
3.3 Selection of B-cell epitopes
We aimed to identify and select B-cell epitopes that target the envelope protein (E) of all four Dengue virus serotypes (DENV1-4). For that purpose, we used Bepipred, Bepipred 2.0, Emini surface accessibility, and Kolaskar & Tongaonkar algorithms from the IEDB database to screen 40 sequences from different serotypes. We chose epitopes that were 8–16 amino acids long and met certain antigenicity and allergenicity thresholds. For DENV1, we found 11 conserved epitopes, 9 with high antigenic scores of 0.4391–1.3954. Three of them—KQEVVVLGS, KYSVIVTVH, and ATEIQTSGTT—had low allergenicity potential, high immunogenicity scores and were conserved in all strains, showing their potential as vaccine targets (Table 3). For DENV2, we found 12 common epitopes, 5 with non-allergenic properties (WQEVVVLGS, VDIVLEHGSCVTT, TEAKQPAT, KQDVVVLGS, and FTGHLKCR) and 7 with high antigenicity (0.4516–1.8735). All the strains fully conserved these epitopes, which supports their potential. We then analysed DENV3 and found 11 common epitopes, 9 with high antigenicity (0.4280–1.5850). Only 5 of them were non-allergenic (VVTKKEEPV, KQEVVVLGS, LATLRKLCIEG, KYTVIITVH, and FVLKKEVSETQH), but they had high immunogenicity scores and were conserved in all strains. Lastly, for DENV4, we found 7 conserved epitopes, but only 2 were not allergic (EMAETQHGT and QGEPYLKEEQDQ). These epitopes had antigenicity values of 0.6967 and 0.5256, respectively, and were found in all the sequences. Overall, this method successfully identified a set of highly conserved, immunogenic, and non-allergenic B-cell epitopes for all four Dengue virus serotypes which were further utilized to design the vaccine candidates.
3.4 Vaccine construction
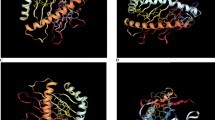
Our vaccine constructs were designed by combining the selected epitopes, and we used appropriate linkers such as AAY, GPGPG, and KK to join the CD4 + and CD8 + T-cell and B-cell epitopes. Various criteria were evaluated to find the optimal vaccine design, including allergenicity, antigenicity, autoimmunity, secondary and tertiary structure quality, and discontinuous epitope scores. By adding several adjuvants with EAAAK linkers, we attempted to improve vaccine quality. Afterwards, we evaluated four designs based on physical–chemical properties, tertiary structure quality (ProSA), toxicity, allergenicity, and antigenicity. DV1 is the original vaccine design, whereas DV2 represents an improved vaccine construct in which 50S ribosomal proteins are attached to the N-terminus and PADRE sequences are attached to the C-terminus. Instead of placing the PADRE sequence at the C-terminus, the DV3 design places it at the N-terminus. Adding the RS09 adjuvant to the C-terminus and the RR motif to the N-terminus of HBD3, the DV4 design boosts the vaccine's effectiveness. In addition to improving vaccine development, the four enhanced 3-D structures in Fig. 4 are listed in Supplementary Table 2.
The predicted 3D models of DV1-4 vaccine models. SubFig A displays the model for DV1 vaccine in red. SubFig B showcases the DV2 vaccine with the 50S ribosomal protein L7/L12 adjuvant highlighted in yellow at the N-terminal and the PADRE sequence in purple at the C-terminal. SubFig C presents the DV3 vaccine model, emphasizing the PADRE sequence in purple at the C-terminal. SubFig D illustrates the DV4 construct, with the HBD3 featuring the RR motif highlighted in pink at the N-terminal and the RS09 adjuvant in green at the C-terminal
3.5 Population coverage
We conducted a global population coverage study to assess the potential of the constructed vaccines to protect against the disease across diverse geographies using the IEDB reference alleles. The results showed that the linked CTL epitopes covered an impressive 98.55% of the world's population, while the combined HTL epitopes achieved a coverage of 81.81%. The multi-epitope vaccine, which included all the selected epitopes, reached an outstanding population coverage of 99.70%, demonstrating its wide-ranging potential for effective vaccination. Supplementary Fig. 1 shows the detailed population coverage data for each region.
3.6 Assessment and validation of designed vaccine models
Then, this study investigated the immunogenicity, biophysical properties, and structural integrity of the constructed vaccines with and without various adjuvants such as 50S ribosomal protein L7/L12, PADRE, HBD3 with the RR motif, and RS09. Our study, as shown in Supplementary Table 3, reveals that none of the formulations were toxic or allergenic. We used the Vaxijen server to predict the antigenicity of the unmodified vaccine (DV1), which scored 0.6718. Interestingly, the antigenicity scores for DV1 were slightly higher than those for the vaccine with any adjuvants, with scores for DV2, DV3, and DV4 at 0.6135, 0.6704, and 0.6392, respectively. We also analyzed the physical characteristics of these vaccine models, which indicated that they had different structural features. The four vaccine models had different chain lengths—DV1 has 215 amino acids, DV2 has 362 amino acids, DV3 has 233 amino acids, and DV4 has 279 amino acids, but all of them demonstrated similar isoelectric point (pI) values, ranging from 9.09 to 9.78. The instability index values indicate that DV1 and DV3 are less stable than DV2 and DV4, which is important to consider. For instance, DV2 has a high aliphatic index, which suggests better thermostability, a desirable feature for long-term transport and storage.
The negative GRAVY scores of these vaccines indicate that they are hydrophilic. In terms of solubility and molecular interactions DV1, DV3, and DV4 all have lower hydrophilicity than DV2. Figure 5 demonstrates that all vaccine models have high-quality protein structures, as most amino acids are in the "most favoured regions," according to our Ramachandran plot analysis. DV4 stands out among the tested vaccines, with 95.6% of its amino acid residues in the "most preferred regions," 4% in additional allowed regions, 0% in generously allowed regions, and 0.4% in disallowed regions. Figure 6 also shows that the Z scores, ranging from -1.58 to -3.55, confirm structural similarity with known protein structures, further enhancing the models' potential for structural biology and vaccine development.
Ramachandran plot analysis for vaccine 3D structure validation of A DV1, B DV2, C DV3, & D DV4 model. Residues in most favored regions [A, B, L] in red regions, Residues in additional allowed regions [a, b, l, p] in yellow regions, Residues in generously allowed regions [~ a, ~ b, ~ l, ~ p] in light grey color regions and white residues in disallowed regions
3.7 Secondary structure prediction
We examined the secondary structures of our four vaccine models with PDBsum, unveiling a captivating landscape of diverse configurations. DV1, adhering to our predictions, displayed a balanced mix of 2 β-strands, 1 β-hairpin, 8 helices, 30 β-turns, and 2 γ-turns (Supplementary Fig. 2). In contrast, DV3 was very different, with no β-hairpins or β-strands, but 11 α-helices, 27 β-turns, and one γ-turn. DV2 and DV4 were very similar, with 2 β-strands, 1 β-hairpin, 14 helices, 36 β-turns, and 3 γ-turns each, suggesting possible commonalities in their function or stability. These intriguing results show the amazing diversity in secondary structure formations among the DV models, each with its own distinctive feature, and set the stage for further investigation of their unique properties and potential in vaccine development.
3.8 Conformational B-cell epitopes
Ellipro server was used to predict discontinuous B cell epitopes of the designed vaccine constructs. The prediction identified 29 vaccine-related epitopes (Supplementary Table 4). These epitopes had scores ranging from 0.509 to 0.934. DV3 was the most productive model, generating nine discontinuous epitopes. DV1 had the fewest conformational epitopes, with only five discontinuous epitopes, due to the lack of adjuvants.
3.9 Molecular docking analysis
Using ClusPro v2.0, we investigate the interaction between vaccination candidates and TLR-4, a key receptor for inducing a strong immune response. Our extensive analysis generated thirty docked complexes for each vaccine construct bound to TLR-4 using this computational method, and then we calculated the centre and lowest energy scores of each and the most promising docked complexes based on the lowest binding energy for each vaccine model were selected (Fig. 7). The most stable configurations with docking score values were − 1150.4, − 1139.1, − 1147.4, and − 1484.5 for DV1, DV2, DV3 and DV4 respectively. Following that, we used PDBsum and ChimeraX to visualize the structural features of the complexes and to understand better the nature of these interactions (Fig. 8A and B).
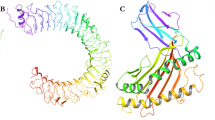
The docked complex of TLR4 and multi-epitope vaccine (DV-4) obtained from ClusPro based on the lowest binding energy. A Depicts the docked complex of TLR4/MD2-vaccine, with the vaccine construct highlighted in red, the TLR4 dimer in green, and MD2 co-receptor in purple. B Highlights the specific interacting residues between the docked TLR4/MD2 tetramer (chain B) and the vaccine (chain E). TLR4 interacting residues are represented in green, MD2 residues in purple sticks, and vaccine contact residues in red spheres (white backbone). C and D Display the residue interactions across the interface, with interacting chains connected by colored lines, each denoting a different type of interaction as indicated in the key provided
Our analysis showed that the TLR-4 receptor (chain B) and the vaccine (chain E) formed a significant interaction network, consisting of 7 salt bridges (between Lys180, Glu27, Glu31, Glu24, Glu42 residues of the receptor & Glu108, Arg46, Arg43, Arg46, lys39, Arg12 of vaccine) and 20 hydrogen bonds (between Asn143, Lys186, Gln163, Gln188, Gln115, Pro28, Cys29, Glu27, Glu31, Glu24, Pro23, Met41, Glu605, Glu603 of receptor residues and Asn131, Ser107, Glu108, Glu124, Arg47, Arg46, Arg 43, Lys39, Arg12, Ala19, Val20, Asn4, Gly1 of vaccine) (Fig. 8C, D). Most of these interactions occurred in the contact area between the two molecules, which covered a large 1544 Å2 for TLR-4 and 1522 Å2 for the vaccine, respectively.
3.10 Immune simulation
To delve deep into the analysis, we used the C-ImmSim server to simulate immune response after the three vaccine doses over a year. The B-cell population demonstrated a significant increase in the presence, production, and levels of IgG1 + IgG2, IgM, and IgG + IgM antibodies during the secondary and tertiary responses as the antigen concentration decreased. The main immune response had IgM and IgM + IgG antibodies. Figure 9A, B, and G show that the IgM and IgM + IgG titers had a clear separation in the first response. The secondary and tertiary responses had a different separation of these peaks. A stronger secondary and tertiary immune response was possible because of a higher total B and TH cell count (Fig. 9B, D). In later exposures, there was an increase in memory cell equivalents, total and active B lymphocytes, and TH cells (Fig. 9C, E, F). There was a lot of IFN during the immune response, especially in the secondary and tertiary responses. This is because 63% of the vaccine's epitopes can produce IFN (Fig. 9H).
3.11 Molecular dynamic simulation
Finally, we used Molecular Dynamics (MD) simulation to study the dynamic behaviour and stability of both the vaccine-TLR-4 complex and the TLR-4 alone, as shown in Fig. 10. Figure 10A shows the initial snapshot of the complex, while Fig. 10B shows the final snapshot, highlighting the effects of different parameters on the complex's behavior.
Molecular Dynamics Simulation of the DV4-TLR-4 Complex (in Blue) and the TLR-4 alone (Gold) at 100 ns: A Initial Snapshot of the Complex; B Post-Simulation Snapshot; C Analysis of Root Mean Square Deviation Highlighting Subtle Structural Dynamics; D RMSF (Root Mean Square Fluctuation) Plot, Demonstrating Side Chain Flexibility; E Evolution of Compactness as Evidenced by Radius of Gyration (Rg); F Solvent Accessible Surface Area (SASA) Demonstrates Stability Throughout the simulation
First of all, The RMSD profiles presented in Fig. 10C provide a dynamic representation of the systems' stability across the simulation period. In both scenarios, an upward trend in RMSD values is evident, denoting conformational adjustments influenced by the vaccine binding to the receptor. Specifically, the RMSD trajectory of the vaccine-receptor complex exhibits an initial fluctuation, starting at 0.50 nm, reaching 1.3 nm after 25 ns, and steadily increasing to 1.70 nm over the 100 ns simulation. In contrast, the receptor alone maintains relatively stable RMSD values throughout. Examining regional flexibility via RMSF analysis (Fig. 10D) reveals pronounced alterations in receptor mobility upon vaccine interaction, particularly in the central protein regions. Notably, residue 600 displays heightened flexibility, with RMSF values ranging from 0.2 to 0.7 nm, implying potential repercussions on interaction dynamics and conformational adaptability. Analysis of radius of gyration (Rg) profiles (Fig. 10E) demonstrates a consistent decrease in both protein configurations throughout the simulation. However, while the receptor maintains an Rg value below 3.25 nm, the vaccine-receptor complex exhibits values fluctuating between approximately 4.00 and 3.25 nm. This reduction in Rg for the complex suggests enhanced stability and ordered conformational changes, indicative of molecular interactions. Furthermore, evaluation of solvent accessible surface area (SASA) dynamics (Fig. 10F) unveils notable disparities between the vaccine-receptor complex and the receptor alone. SASA values for the complex remain relatively constant, surpassing 500 nm2, indicating sustained exposure to solvents and inherent flexibility. In contrast, the receptor displays lower SASA values, typically below 400 nm2, signifying limited exposure of its hydrophobic core to solvents.
3.12 Codon optimization and in-silico cloning
To achieve in silico cloning, vaccines must be expressed efficiently into an E. Coli expression system. The expected epitopes for T-cells and B-cells, along with the proper adjuvants and linkers, were used to construct four vaccines (DV-1–4). The Java codon adaptation tool (JCAT) was utilized to modify the codon usage to accommodate the majority of sequenced prokaryotic organisms using the sequences of these vaccines as input (individually). According to the analysis's findings, the DNA sequences of DV-1–4 were, in order, 645, 1086, 699, and 837 nucleotides. The observed CAIs (DV-1 = 0.98; DV-2 = 0.98; DV-3 = 0.98 and DV-4 = 0.99) indicated that the adapted sequences were made up of codons capable of cellular machinery of the target organism. Furthermore, the optimized sequences had a GC content between 46 and 49%. The sequence data suggests that the proposed vaccines are reliably expressed and of high efficiency in E. coli. Restriction recognition sites of the restriction enzymes XhoI and BstAPI were conjugated at the N and C-terminal of reverse translated nucleotides of DV-1 and 3 respectively and XhoI and MluI were conjugated at the N and C terminal of reverse translated nucleotides of DV-2 and 4 respectively (Fig. 11) and the final optimized sequences were inserted into the pET28a ( +) vector in order to clone the designed vaccines using SnapGene software. The cloned plasmids for DV-1–4 had final lengths of 5378, 5496, 5432, and 5247 kbp, in that order (Fig. 12).
In Silico Restriction Cloning of Codon-Optimized Multi-Epitope Vaccine sequences highlighted A DV-1 in Red, B DV-2 in Purple, C DV-3 in Green, and D DV-4 in Blue into a pET-28a( +) Vector colored in Black. Restriction sites XhoI, BstAPI (for DV-1 and DV-3), and XhoI, MluI (for DV-2 and DV-4) are conjugated at the N and C-terminal regions of the vaccines, allowing for efficient cloning. The final constructs are suitable for expression in E. coli (strain K12) for vaccine production
4 Discussion
Over 100 countries are endemic to dengue, a global health problem since 1779 [47]. Over the last 50 years, dengue incidence has increased 30-fold due to various complex factors [48] (CDC 2014). Vaccination is an important strategy to control these high-prevalence diseases by reducing the disease burden and preventing virus transmission [49]. Ineffective treatments and vaccines, however, have worsened the dengue situation [50, 51]. A tetravalent vaccine is necessary for complete protection against dengue disease; the high mutation rates of the serotypes make developing a dengue vaccine challenging [52, 53]. A vaccine against dengue has been developed by Sanofi Pasteur and is marketed as Dengvaxia®. There is, however, a difference in the vaccine's efficacy depending on the serotype and the region, as well as the fact that the vaccine is only administered to seropositive individuals [54].
The development of a potent tetravalent dengue vaccine has been attempted numerous times [11, 12, 14, 55], but these efforts have failed to elicit a sufficient immune response against one of the serotypes, mainly DENV 4 [56]. To develop a successful dengue vaccine, the vaccine must induce immunity against all four serotypes, regardless of location. To accomplish this, we targeted conserved structural and non-structural proteins from all four serotypes. Based on the conservation of the virus proteins and their ability to induce a sufficient immune response when vaccinated, we selected E, NS1, NS3, and NS5 for our analysis. There have been previous vaccine approaches to the dengue virus using the E protein [57], the NS1 protein [58], the NS3 protein [59], and the NS5 protein [60]. However, to our knowledge, none of these four proteins have been combined with epitopes from all the proteins to construct a vaccine.
In this study, we conducted a comprehensive bioinformatics analysis to design a possible dengue vaccine, which included retrieving polypeptide sequences, phylogenetic analysis, epitope prediction, designing vaccine candidates, molecular docking of TLR-4 and the vaccine construct, dynamic simulation of the complex using GROMACS (version 2020.6), cloning into a vector and immune simulation using C-ImmSim. Our results provide valuable insights to develop an effective dengue vaccine. For instance, the phylogenetic analysis of 40 polypeptide sequences from Dengue virus serotypes 1 to 4 revealed complex patterns of genetic relatedness. Despite their geographical distance, dengue virus strains had close genetic similarities, suggesting a complex genetic evolution and transmission dynamics [61]. The diversity and evolution of dengue virus strains underscores the need for a comprehensive vaccine strategy.
In our epitope prediction analysis of CD4 + , CD8 + T-cells, and B-cells, we identified thousands of unique epitopes but selected only those common to all strains. These epitopes were found to have strong antigenic properties, were non-allergenic, and were non-toxic, making them promising vaccine candidates. Vaccine candidates were constructed by joining CD4 + and CD8 + T-cell and B-cell epitopes with suitable linkers. Our four vaccine candidates (DV1, DV2, DV3, and DV4) were created using different strategies, each with unique features and adjuvants, to improve the vaccine's potential [62,63,64]. An important aspect of our dengue vaccine candidate is that the population coverage analysis showed that the vaccine could cover 99.70% of the global population. The presence of discontinuous B-cell epitopes in all models further illustrates the vaccine's potential by targeting different aspects of the immune response [65].
The secondary structure analysis revealed variations in the composition of secondary structure elements across the vaccine models, with some models featuring β-strands, helices, and β-turns, while α-helices and β-turns characterized others. The structural differences may have an impact on the immune response as well as the stability of proteins. Compared to previous vaccine design studies [28, 62, 66, 67], these vaccine models were found to be of high quality, antigenic, and safe, although some were more thermostable and hydrophobic than others. According to the antigenicity analysis, one vaccine model without an adjuvant had the highest antigenicity, which is atypical. Therefore, this model may induce a powerful immune response without extra adjuvants, which could be extremely useful for vaccine development.
Further, molecular docking analysis revealed positive interactions with the TLR-4 receptor, an essential immune system component [68]. A strong interaction network was observed between vaccine models and TLR-4 receptors through salt bridges and hydrogen bonds. It was apparent that the vaccine and immune receptor had a high interaction surface area, which suggests a high affinity for binding [67], crucial for triggering a robust immune response. During the interaction between the vaccine and the receptor, the molecular dynamics simulation results showed dynamic structural changes, changes in receptor mobility, increased system compactness, and constant solvent exposure [63].Overall, the attachment of the vaccine molecule to the receptor seemed to alter the toll-like receptor, causing it to become more flexible, unfurl, and become more accessible to solvents, as compared to its stable form without the attached molecule. The changes in the receptor caused by the vaccination at a molecular level give us information about the structural changes and movements that occur during receptor activation and subsequent signaling. The receptor's flexibility and unfolding are enhanced when the vaccine binds to it, indicating that the receptor may assume a "activated '' open conformation. This conformation could potentially assist subsequent contacts and signaling processes. Additional pathway analysis and mutational investigations are required to validate; however, the vaccine-induced instability and conformational shifts of the receptor indicate that these structural changes may contribute to the activation of downstream signaling.. By analyzing these interactions, we can better understand how proteins interact with their ligands and how this affects their structural dynamics. Last but not least, the immune simulation graphs demonstrate that the immune system recognized and responded effectively to the virus, eliminating it and producing antibodies in the process. It is based on the growth of active T and B cells after simulated antigen injections. Lack of immune tolerance or suppression, low levels of anergic cells, and the presence of key molecules indicate a robust immune response that produces antibodies to clear the infection [69, 70].
Therefore, our study contributes significantly to the development of a much-needed vaccine for dengue. Our findings provide a solid foundation for further research and development efforts, although the virus remains challenging from a genetic, structural, and immune perspective.
5 Conclusion
In conclusion, this study designed a multi-epitope vaccine candidate using bioinformatics to fight the dengue virus, a global health threat. Following established vaccine development principles, we identified and selected highly immunogenic and conserved epitopes targeting structural and nonstructural dengue proteins (E, NS1, NS3, and NS5) from all four serotypes [1,2,3,4]. By assessing its allergenicity, antigenicity, and ability to stimulate humoral and cellular immune responses, we ensured broad population coverage, reaching 99.70% of the global population, while prioritizing safety and efficacy. Importantly, our analysis using a Ramachandran plot revealed that 95.6% of the amino acid residues of the vaccine candidates reside within the optimal zone, with around 4% in additional allowed regions and 0.4% in disallowed regions. Different adjuvants (50S ribosomal protein L7/L12, RS09, HBD3, PADRE sequences) were used to construct and optimize four different vaccine models. Molecular dynamics simulations were then used to analyze the molecules' structural integrity, interactions with receptors, and dynamic behavior. An agent-based modelling server was used to simulate the immune response to each vaccine model to gain a deeper understanding of immunogenicity. These findings indicate the potential of these multi-epitope vaccine candidates. There is still much work to be done to validate their efficacy in combating dengue virus infection, leading to the development of an effective and novel vaccine against this widespread global health threat.
Data of availability and materials
Data is contained within the article and Supplementary Material.
References
Liu SY, Chien TW, Yang TY, Yeh YT, Chou W, Chow JC. A bibliometric analysis on dengue outbreaks in tropical and sub-tropical climates worldwide since 1950. Int J Environ Res Public Health. 2021;18(6):3197.
WHO. Dengue and severe dengue [Internet]. 2023 [cited 2023 Nov 10]. Available from: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue
Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, et al. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl Trop Dis. 2012;6(8):e1760.
Bhatnagar P, Sreekanth GP, Murali-Krishna K, Chandele A, Sitaraman R. Dengue virus non-structural protein 5 as a versatile, multi-functional effector in host-pathogen interactions. Front Cell Infect Microbiol. 2021;18:11.
Płaszczyca A, Scaturro P, Neufeldt CJ, Cortese M, Cerikan B, Ferla S, et al. A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle. PLoS Pathog. 2019;15(5):e1007736.
Chen HR, Lai YC, Yeh TM. Dengue virus non-structural protein 1: a pathogenic factor, therapeutic target, and vaccine candidate. J Biomed Sci. 2018;25(1):58.
Thomas SJ, Martinez LJ, Endy TP. Flaviviruses: Yellow Fever, Japanese B, West Nile, and Others. In: Viral Infections of Humans. Boston, MA: Springer US; 2014. pp. 383–415.
Halstead SB. Identifying protective dengue vaccines: guide to mastering an empirical process. Vaccine. 2013;31(41):4501–7.
Simmons JS, St John JH, Reynolds FHK. Experimental studies of dengue. Philippine J Sci. 1931;44(1–2):1–251.
Ratto-Kim S, Yoon IK, Paris RM, Excler JL, Kim JH, O’Connell RJ. The US military commitment to vaccine development: a century of successes and challenges. Front Immunol. 2018;21:9.
Danko JR, Kochel T, Teneza-Mora N, Luke TC, Raviprakash K, Sun P, et al. Safety and immunogenicity of a tetravalent dengue DNA vaccine administered with a cationic lipid-based adjuvant in a phase 1 clinical trial. Am J Trop Med Hyg. 2018;98(3):849–56.
Whitehead SS, Falgout B, Hanley KA, Blaney Joseph E, Markoff L, Murphy BR. A live, attenuated dengue virus type 1 vaccine candidate with a 30-nucleotide deletion in the 3′ untranslated region is highly attenuated and immunogenic in monkeys. J Virol. 2003;77(2):1653–7.
Salje H, Alera MT, Chua MN, Hunsawong T, Ellison D, Srikiatkhachorn A, et al. Evaluation of the extended efficacy of the Dengvaxia vaccine against symptomatic and subclinical dengue infection. Nat Med. 2021;27(8):1395–400.
Kirkpatrick BD, Whitehead SS, Pierce KK, Tibery CM, Grier PL, Hynes NA, et al. The live attenuated dengue vaccine TV003 elicits complete protection against dengue in a human challenge model. Sci Transl Med. 2016;8(330):330ra36.
Brisse M, Vrba SM, Kirk N, Liang Y, Ly H. Emerging concepts and technologies in vaccine development. Front Immunol. 2020;30:11.
Fadaka AO, Sibuyi NRS, Martin DR, Goboza M, Klein A, Madiehe AM, et al. Immunoinformatics design of a novel epitope-based vaccine candidate against dengue virus. Sci Rep. 2021;11(1):19707.
Lim SP. Dengue drug discovery: progress, challenges and outlook. Antiviral Res. 2019;163:156–78.
Guzman MG, Halstead SB, Artsob H, Buchy P, Farrar J, Gubler DJ, et al. Dengue: a continuing global threat. Nat Rev Microbiol. 2010;8(S12):S7-16.
Halstead SB. Dengue hemorrhagic fever: two infections and antibody dependent enhancement, a brief history and personal memoir. Rev Cubana Med Trop. 2002;54(3):171–9.
Shukla R, Ramasamy V, Shanmugam RK, Ahuja R, Khanna N. Antibody-dependent enhancement: a challenge for developing a safe dengue vaccine. Front Cell Infect Microbiol. 2020;22:10.
Murphy BR, Whitehead SS. Immune response to dengue virus and prospects for a vaccine. Annu Rev Immunol. 2011;29(1):587–619.
Chauhan V, Rungta T, Goyal K, Singh MP. Designing a multi-epitope based vaccine to combat Kaposi Sarcoma utilizing immunoinformatics approach. Sci Rep. 2019;9(1):2517.
Zhang L. Multi-epitope vaccines: a promising strategy against tumors and viral infections. Cell Mol Immunol. 2018;15(2):182–4.
Mahmud S, Rafi MO, Paul GK, Promi MM, Shimu MSS, Biswas S, et al. Designing a multi-epitope vaccine candidate to combat MERS-CoV by employing an immunoinformatics approach. Sci Rep. 2021;11(1):15431.
Khairkhah N, Bolhassani A, Agi E, Namvar A, Nikyar A. Immunological investigation of a multiepitope peptide vaccine candidate based on main proteins of SARS-CoV-2 pathogen. PLoS ONE. 2022;17(6):e0268251.
Yang Z, Bogdan P, Nazarian S. An in silico deep learning approach to multi-epitope vaccine design: a SARS-CoV-2 case study. Sci Rep. 2021;11(1):3238.
Ma J, Qiu J, Wang S, Ji Q, Xu D, Wang H, et al. A novel design of multi-epitope vaccine against helicobacter pylori by immunoinformatics approach. Int J Pept Res Ther. 2021;27(2):1027–42.
Bhardwaj A, Sharma R, Grover A. Immuno-informatics guided designing of a multi-epitope vaccine against Dengue and Zika. J Biomol Struct Dyn. 2023;41(1):1–15.
Sabetian S, Nezafat N, Dorosti H, Zarei M, Ghasemi Y. Exploring dengue proteome to design an effective epitope-based vaccine against dengue virus. J Biomol Struct Dyn. 2019;37(10):2546–63.
Ali M, Pandey RK, Khatoon N, Narula A, Mishra A, Prajapati VK. Exploring dengue genome to construct a multi-epitope based subunit vaccine by utilizing immunoinformatics approach to battle against dengue infection. Sci Rep. 2017;7(1):9232.
Koichiro Tamura, Glen Stecher, Sudhir Kumar. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Molecular Biology and Evolution; 2021. p. 3022–7.
Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43(1):D405–12.
Ponomarenko J, Bui HH, Li W, Fusseder N, Bourne PE, Sette A, et al. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008;9(1):514.
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, et al. Protein Identification and Analysis Tools on the ExPASy Server. In: Handbook TPP, editor., et al., Totowa. NJ: Humana Press; 2005. p. 571–607.
Sharma N, Naorem LD, Jain S, Raghava GPS. ToxinPred2: an improved method for predicting toxicity of proteins. Brief Bioinform. 2022;23(5):bbac174.
Doytchinova IA, Flower DR. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007;8(1):4.
Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP vol 2—a server for in silico prediction of allergens. J Mol Model. 2014;20(6):2278.
Cheng J, Randall AZ, Sweredoski MJ, Baldi P. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005;33(Web Server):W72–6.
Meng EC GTPECGPZMJF TE. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci; 2023. p. e4792.
Heo L, Park H, Seok C. GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013;41(W1):W384–8.
Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server):W407–10.
Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, et al. The ClusPro web server for protein–protein docking. Nat Protoc. 2017;12(2):255–78.
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25.
Rapin N, Lund O, Castiglione F. Immune system simulation online. Bioinformatics. 2011;27(14):2013–4.
Morla S, Makhija A, Kumar S. Synonymous codon usage pattern in glycoprotein gene of rabies virus. Gene. 2016;584(1):1–6.
Kamens J. The Addgene repository: an international nonprofit plasmid and data resource. Nucleic Acids Res. 2015;43(D1):D1152–7.
Diamond MS, Pierson TC. Molecular insight into dengue virus pathogenesis and its implications for disease control. Cell. 2015;162(3):488–92.
Ebi KL, Nealon J. Dengue in a changing climate. Environ Res. 2016;151:115–23.
Rose N, Andraud M. The use of vaccines to control pathogen spread in pig populations. Porcine Health Manag. 2017;3(1):8.
Cooper S, Schmidt BM, Sambala EZ, Swartz A, Colvin CJ, Leon N, et al. Factors that influence parents’ and informal caregivers’ views and practices regarding routine childhood vaccination: a qualitative evidence synthesis. Cochrane Database Syst Rev. 2021;2021(10):CD013265.
Chawla P, Yadav A, Chawla V. Clinical implications and treatment of dengue. Asian Pac J Trop Med. 2014;7(3):169–78.
Dolan PT, Taguwa S, Rangel MA, Acevedo A, Hagai T, Andino R, et al. Principles of dengue virus evolvability derived from genotype-fitness maps in human and mosquito cells. Elife. 2021;25:10.
Raviprakash K, Wang D, Ewing D, Holman DH, Block K, Woraratanadharm J, et al. A tetravalent dengue vaccine based on a complex adenovirus vector provides significant protection in rhesus monkeys against all four serotypes of dengue virus. J Virol. 2008;82(14):6927–34.
Torresi J, Ebert G, Pellegrini M. Vaccines licensed and in clinical trials for the prevention of dengue. Hum Vaccin Immunother. 2017;13(5):1059–72.
Roy SK, Bhattacharjee S. Dengue virus: epidemiology, biology, and disease aetiology. Can J Microbiol. 2021;67(10):687–702.
Biswal S, Reynales H, Saez-Llorens X, Lopez P, Borja-Tabora C, Kosalaraksa P, et al. Efficacy of a tetravalent dengue vaccine in healthy children and adolescents. N Engl J Med. 2019;381(21):2009–19.
Metz SW, Tian S, Hoekstra G, Yi X, Stone M, Horvath K, et al. Precisely molded nanoparticle displaying DENV-E proteins induces robust serotype-specific neutralizing antibody responses. PLoS Negl Trop Dis. 2016;10(10):e0005071.
Beatty PR, Puerta-Guardo H, Killingbeck SS, Glasner DR, Hopkins K, Harris E. Dengue virus NS1 triggers endothelial permeability and vascular leak that is prevented by NS1 vaccination. Sci Transl Med. 2015; 7(304).
Costa SM, Yorio AP, Gonçalves AJS, Vidale MM, Costa ECB, Mohana-Borges R, et al. Induction of a protective response in mice by the dengue virus NS3 protein using DNA vaccines. PLoS ONE. 2011;6(10):e25685.
Lim SP, Noble CG, Shi PY. The dengue virus NS5 protein as a target for drug discovery. Antiviral Res. 2015;119:57–67.
Worobey M, Holmes EC. Evolutionary aspects of recombination in RNA viruses. J Gen Virol. 1999;80(10):2535–43.
Mahmoodi S, Amirzakaria JZ, Ghasemian A. In silico design and validation of a novel multi-epitope vaccine candidate against structural proteins of Chikungunya virus using comprehensive immunoinformatics analyses. PLoS ONE. 2023;18(5):e0285177.
Salaikumaran MR, Kasamuthu PS, Aathmanathan VS, Burra VLSP. An in silico approach to study the role of epitope order in the multi-epitope-based peptide (MEBP) vaccine design. Sci Rep. 2022;12(1):12584.
Singh A, Thakur M, Sharma LK, Chandra K. Designing a multi-epitope peptide based vaccine against SARS-CoV-2. Sci Rep. 2020;10(1):16219.
Yurina V, Adianingsih OR. Predicting epitopes for vaccine development using bioinformatics tools. Ther Adv Vaccines Immunother. 2022;21(10):251513552211002.
Omoniyi AA, Adebisi SS, Musa SA, Nzalak JO, Bauchi ZM, Bako KW, et al. In silico design and analyses of a multi-epitope vaccine against Crimean-Congo hemorrhagic fever virus through reverse vaccinology and immunoinformatics approaches. Sci Rep. 2022;12(1):8736.
Kaushik V, G SK, Gupta LR, Kalra U, Shaikh AR, Cavallo L, et al. Immunoinformatics Aided Design and In-Vivo Validation of a Cross-Reactive Peptide Based Multi-Epitope Vaccine Targeting Multiple Serotypes of Dengue Virus. Front Immunol. 2022;13.
Zhang Y, Liang X, Bao X, Xiao W, Chen G. Toll-like receptor 4 (TLR4) inhibitors: Current research and prospective. Eur J Med Chem. 2022;235:114291.
Rocamora-Reverte L, Melzer FL, Würzner R, Weinberger B. The complex role of regulatory T cells in immunity and aging. Front Immunol. 2021;27:11.
Sun L, Su Y, Jiao A, Wang X, Zhang B. T cells in health and disease. Signal Transduct Target Ther. 2023;8(1):235.
Acknowledgements
The authors acknowledge the Noakhali Science and Technology University Research Cell (NSTURC) and R & D Project, Ministry of Science and Technology, Bangladesh for providing the support to the researchers.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Author information
Authors and Affiliations
Contributions
Conceptualization OS, and AR; Data curation OS, AR, NS, and NR; Formal analysis AR, NR, and ABZ; Investigation OS, AR and NS; Methodology OS, AR, AS, TAS, and FH; Resources OS, MMR, and NMB; Software OS, AR and MRA; Supervision OS, NS, and MSA; Validation NR, AR, AS, MSA and MMR; Visualization AR, and NR; Writing -original draft OS, AR, NR and ABZ; Writing—review & editing OS, NS, AS, NMB, MMR, and FH. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Saha, O., Razzak, A., Sarker, N. et al. In silico design and evaluation of multi-epitope dengue virus vaccines: a promising approach to combat global dengue burden. Discov Appl Sci 6, 210 (2024). https://doi.org/10.1007/s42452-024-05782-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-024-05782-9