Abstract
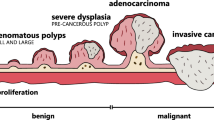
This study describes an automated detection of polyp type as it is very important to determine the existence of dysplasia—a stage leading to the development of gastrointestinal cancer. The polyp-type classification is performed by a multiclass support vector machine from feature-fusion of bi-dimensional empirical mode decomposition (BEMD) and convolutional neural network (CNN). An extensive experiment is performed using standard datasets by extracted features from the individual technique as well as a fusion of features from BEMD and CNN. The fusion technique confirms satisfactory performance compared to other techniques with an accuracy of 98.94%. Moreover, it shows potentiality in precisely classifying some challenging polyps even though these are somehow confusing for human experts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
According to the world health organization, cancer is becoming the most leading cause of death around the world. Gastrointestinal cancer is very dreadful among them. The American Cancer Society has published a statistic in 2020 that 53,200 cancer-related deaths are predicted from 147,950 cases of gastrointestinal cancers [1]. In the USA, the statistic showed that it stood third among the most pervasive diseases and the second-largest cause of death in cancer [1]. However, it can be curable at an early stage if the malignancy of polyps is detected and classified at the preliminary level.
The usual medical method for gastrointestinal polyp diagnosis is video endoscopy. Polyp’s shape and type are very confusing to the operators as these depend on various parameters. Sometimes the misclassification causes severe malignancy in the later stage. But all the categories of polyp are not exhibiting the same risk. Therefore, accurate polyp classification is very crucial.
Clinical studies have found gastrointestinal polyp as the precursor of developing cancer cells [2,3,4,5]. Several diagnosis criteria including routine endoscopy have been introduced to help the medical doctors in polyp detection. Usually, a polyp can be classified as Lumen, Diverticula, Adenoma, Hyperplastic, Serrated. The operator-dependent procedure of endoscopic examination may lead to polyp misdetection as well as misclassification. To reduce this error, the computer-aided system is very much helpful.
Recently, computer-aided polyp detection systems have been developed to work on endoscopic video frames using color, shape, texture, and spatial features [6,7,8]. These help in the diagnosis process of polyp detection, and, also indicates the affected regions that might be examined with more attention [9, 10]. In [11], image feature extraction from the covariance matrix of textural behavior of wavelet decomposition and linear discriminant analysis (LDA) was proposed for colonic tumor detection. An intelligent system was proposed using the combination of a support vector machine (SVM) and color-texture. It achieved around 94% accuracy. Vassilis and Maria [12] proposed an adaptive neuro-fuzzy system that can extract texture spectra in video capsule endoscopy (VCE) for polyp detection. A comparison of texture-based and color-based methods of polyp detection is investigated by Alexandre et al. [13]. Li et al. [14] reported a classifier using a multilayer perceptron along with SVM for polyp detection using color and shape features and achieved about 94% accuracy. Tajbakhsh et al. [15] proposed an automated polyp detection technique using global geometric constraints and local intensity variations. Our group reported two endoscopic video-based polyp detection techniques: one using color wavelet and convolutional neural network (CNN) features [16] and the other using bi-dimensional empirical mode decomposition (BEMD) and CNN features [17]. Now, it is time to concentrate on the polyp classification [18, 19]. Hence, the main focus of this paper is endoscopic video-based gastrointestinal polyp classification by SVM using the fusion of two kinds of features: BEMD and CNN.
The major contributions of this paper are:
-
i.
We develop an efficient polyp classification through the fusion of BEMD and CNN features.
-
ii.
We perform a detailed experiment to confirm the effectiveness of the proposed technique and compared it with other existing techniques.
-
iii.
We overcome the difficulties of diagnostic experts to identify the different classes of polyps through an automated system with improved accuracy.
The remaining part of this paper is structured as follows:
Section 2 describes the proposed methodology. Polyp region segmentation as well as identification is highlighted in Sect. 3. The experimental results are analyzed in Sect. 4. Finally, conclusions are drawn in Sect. 5.
2 Methodology
This section outlines the proposed experiment to classify polyp classes. The system takes an endoscopic video as input and classifies the polyp in one of the five classes in the output. At first, frames are extracted from the video. Then after enhancement of each frame, a sliding window moves on the frame to capture image patches. After that, feature extractors run on the collected patches to grasp features using BEMD and CNN techniques. A final vector for a polyp classifier is prepared using the fusion of BEMD and CNN features. The SVM has performed to detect the polyp classes in the detection scheme. The self-explanatory major steps of the methodology are given in Fig. 1.
2.1 Dataset
In this paper, we have used 250 videos from two standard publicly available datasets: Endoscopic Vision Challenge dataset (https://polyp.grand-challenge.org/databases/) [20] and Department of Electronics, University of Alcala dataset (http://www.depeca.uah.es/colonoscopy_dataset/) [21]. From these 250 videos, we have extracted 14,900 frames with 5 different polyp types. Lumen and Diverticula type-polyp frames are extracted from Ref. [20] and Adenoma, Hyperplastic, and Serrated are from Ref. [21]. The detailed distributions of extracted video frames are given in Table 1.
2.2 Endoscopic video preprocessing
As mentioned earlier, the experiment has performed on 250 different endoscopic videos. In the training phase when any video feeds into the proposed system, then it transforms into the corresponding frames. A preprocessing technique is required to eliminate the unnecessary parts and irrelevant regions of the separated frames. Some of the extracted frames of different polyp classes are shown in Fig. 2.
Each frame is divided into sub-images of patch size 227 × 227 as shown in Fig. 3. The patch window goes from the top-left region to the bottom-right region of each frame with a stride of 15 × 15. Through these patch collections, a sufficient amount of data has been produced from the captured frames. As supervised learning, this experiment demands huge quantities of training data to train the model. For the training scheme, it has required a sufficient number of the labeled dataset. Those image patches have alleviated the inadequacy of the labeled image. BEMD and CNN features are taken from these image patches.
Sub-images (of size 227 × 227 pixels) inside a frame that is taken from Ref. [20]
2.3 Feature extraction
This section describes the feature extraction process using the proposed feature extractors: Bi-dimensional empirical mode decomposition (BEMD) and Convolutional Neural Network (CNN). The following Sects. 2.3.1 and 2.3.2 describe the BEMD and CNN feature extractors briefly.
2.3.1 Bi-dimensional empirical mode decomposition (BEMD)
Empirical mode decomposition (EMD) is a signal decomposition method, mostly used in signal processing to analyze the intuitive signal decomposition. In this research, we have used the self-adapting power of EMD in 2D image processing named bi-dimensional empirical mode decomposition (BEMD). BEMD, a 2D extension of EMD, has been developed for multiresolution image decomposition purposes [22,23,24,25]. It considers the 2D images as a series of bi-dimensional intrinsic mode functions (BIMFs) and a 2D residual with the same dimension of the original image. This process obeys two basic conditions:
-
The number of extrema in the complete dataset must be equal to the number of zero crossings.
-
The mean value of the local maxima and minima at any point of IMF should be zero.
Image decomposition into IMFs follows a shifting algorithm [17]. If the input image window is denoted as f(x, y), the proposed algorithm can be described as,

Executing this algorithm, a set of BIMFs and a residual r are acquired. They can reflect the original image as
Here, full decomposition of 2D multispectral image has been performed using BEMD. The proposed method separates each RGB image into three color channels (Red, Green, Blue). From there, take the first five BIMFs for analysis as textures, shapes, edges among 9-level BEMD features. Hence, twenty multi-scale images for each channel in four different directions: 0°, 45°, 90°, and 135° are found. This work computes thirteen different features containing statistical and GLCM features for each image, hence, a total of 780 BEMD features for three color channels. If the total pixels in the non-polyp region are defined as M and total pixels in the polyp region is N while the gray-level-color-intensity in polyp region is represented as GI for each pixel, then the statistical features are calculated using Eqs. (2)–(9) as follows.
Along with the statistical features, the calculation of GLCM features has a great value on the textural feature extraction from images. The five GLCM features used in this work are represented using Eqs. (10)–(14).
where the dimension of the image is M × N and \(P\left( {i,j} \right)\) is the absolute value of intensity change of the pixel compared to its previous pixel.
2.3.2 Convolutional neural network (CNN)
CNN outperforms usual state-of-the-art feature extraction methodologies in recent years [26]. Experiment on different CNN models, such as AlexNet, VGG16, VGG19, ResNet50, confirms that the AlexNet model performs better in our dataset. The convolution parts of the AlexNet architecture have extracted 4096 CNN features for each sub-image of size 227 × 227 in 3 color channels. This model has used five convolutional layers followed by a ReLU, 3 max-pooling layers, and 3-fully-connected layers. As a five-class classification scheme, this model has used 5 neurons in the output layer. The dropout functionality has been applied in the fully-connected layers with a 70% probability implementation. Along with dropout and an L2 regularization is remarkably effective to reduce overfitting as well as welfare for generalization error. The ‘softmax’ activation has applied for output which was later removed by the classifier. The architecture of this model is given in Table 2.
2.3.3 Support vector machine (SVM)
Detection and classification are usually done by using SVM [27,28,29,30], neural networks [16, 17, 31, 32], linear discriminant analysis (LDA) [11], adaptive neuro-fuzzy inference system [12], etc. Here, we used the SVM classifier, as it is robust for noisy and sparse data [29]. 780 color EMD features and 4096 CNN features are combined to construct the SVM input vector of 4876 features.
In this research, the classification layer of CNN is replaced by a multiclass SVM, as it uses less epoch and less processing time than the CNN classifier. SVM mimics the principle of one-against-all classification. A hyperplane is considered that will divide each class with all other classes. This hyperplane tries to fit as the maximum distance between the data points of two halves. In this scheme, the SVM is interested to solve Ln binary classification where each current class Li is classified versus other Ln−1 class data. For the total of y data points, SVM will get Ly weight vectors as W1, W2,…, WLy. When trying to classify any new point (x*), it will impose in a position further from the decision boundary by using h(x*). The class can be set by the following optimization equation.
Figure 4 depicts the principle of SVM for five classes in this research.
3 Polyp region identification
After classifying polyp, this work has developed a procedure to identify and mark the polyp regions. Whenever any patch is detected as a polyp, the polyp region is marked by a square. A new circle is formed covering the most overlapped squares.
There are many possible polyp regions of various sizes. Figure 5a shows such possible polyp regions (see the marked regions). The classifier evaluates each marked region by a score. The final polyp region is extracted by averaging the higher score regions (see Fig. 5b). The detected polyp is classified as Hyperplastic.
Polyp detection and classification: a several regions are marked as the candidate for a possible polyp, b a recognized polyp (marked region) and it is classified as Hyperplastic in a video frame taken from Ref. [20]
4 Experimental results and discussions
As mentioned in Sect. 2.1, we have done experimentation using 14,900 frames with different polyp types taken from two publicly available standard datasets. This experiment splits the whole dataset into two groups: 70% data (9900 polyp frames) are for training and 30% data (5000 polyp frames) are for testing. The training features are extracted from the training data and used to train the classifier. After that, testing is performed using testing data and classification performance is evaluated through the well-known metrics derived from the confusion matrix: accuracy, sensitivity, specificity, precision, F-value. These can be presented by the following equations.
where TP is the true positive, FP is the false positive, TN is the true negative, and FN is the false negative. These metrics are performed on different image transform techniques based on GLCM and statistical features.
The first experimentation is done using only BEMD features. The classification results are shown in Table 3. In this table, we have also shown the obtained results from the direct original image features and the wavelet decomposition method for comparison. Performance figures are calculated using the average of all the polyp classes. The results confirm that the BEMD feature-based method is more effective.
The second experimentation is done with various architectural models of CNN. From Table 4, it is confirmed that AlexNet gives better accuracy compared to VGG16, VGG19, and ResNet50 models. For this, in the feature fusion, we used AlexNet model. Moreover, instead of SVM, we have also tried to classify the polyps using CNN. We have found that the SVM classifier is more efficient than the CNN classifier in terms of computation time and accuracy.
The third experimentation is done with feature fusion of BEMD and CNN to classify five types of polyps: Lumen, Diverticula, Adenoma, Hyperplastic, Serrated. Table 5 shows the confusion matrix for each type of polyp classification. The system faces no difficulty in classifying hyperplastic polyp as it is clear enough and quickly detect using the learning features. In a few instances, Lumen and Diverticula look similar and thus making the classification confusing. Similar phenomena are occurring for Serrated and Adenoma. Table 6 shows the performance measure (accuracy, sensitivity, specificity, precision, and F-measure) for the classification of different polyp types. In this table, for comparison purposes, we have also shown the obtained results using CNN only features in classifying different polyp types. The results confirm that our proposed fusion method performs nicely compared to the CNN only feature-based method.
Finally, we have observed the performance of k-fold (k = 5) cross-validation. The whole dataset has randomly partitioned into k equal subsamples. Among the k sub-samples, all the k − 1 subsamples data are considered for the training set and only the remaining subsample has been used for testing the model. This process has repeated for k times by considering each subsample of k once for validating while all other k − 1 is for training. Table 7 shows the 5-fold cross-validation results for BEMD, CNN, and the fusion of BEMD and CNN. The results clearly show that the proposed fusion method outperforms both BEMD only and CNN only feature-based methods.
A few works have been performed on endoscopic polyp classification due to data scarcity. Zou et al. [33] used DCNN in colonoscopy polyp classification. Experimenting with our data set we obtain 94% accuracy for DCNN. Pablo Mesejo et al. [21] used the computer-aided classification for Hyperplastic, Serrated, and Adenoma polyps. They analyzed texture and color features and used a multi-SVM as a classifier and obtained 90% accuracy. The study of Ribeiro et al. [18] classified only two polyp classes—Hyperplastic and Adenoma using the CNN model. They obtained 90.96% accuracy. The average accuracy in polyp classification using the proposed technique is compared with other polyp detection techniques and is shown in Table 8. The table confirms that our proposed fusion-based method shows improved accuracy than the existing polyp detection techniques.
Figure 6 shows some instances where a human expert faces difficulty, but these are correctly classified by our proposed method. Hence our automated system helps human radiological experts.
5 Conclusions
Machine-based polyp classification helps the physicians precisely diagnosis of gastrointestinal cancer at the early stage of polyps. With this motive, here we investigated an automated system for gastrointestinal polyp classification by SVM using the feature-fusion of BEMD and CNN techniques. We did three vital experiments using standard datasets to classify the polyps into five classes. In the first experiment, we have found that BEMD feature-based method is effective than the wavelet decomposition technique. In the second experiment, we have found that AlexNet gives better accuracy compared to VGG16, VGG19, and ResNet50 CNN models for our dataset. In the third experiment, we have performed an in-depth performance analysis of polyp classification using our BEMD and AlexNet CNN feature-fusion method. We have found that our fusion-based polyp classification method shows promising performance (accuracy over 98%) than the individual feature-based method as well as other existing polyp detection techniques. In the future, we will do more detailed experiments and try to fusion other important features for further improvement of accuracy to avoid misclassification for real-life implementation.
References
American Cancer Society. https://www.cancer.org/cancer/colon-rectal-cancer/about/key-statistics.html. Accessed 18 Mar 2020
Park SY, Sargent D (2016) Colonoscopic polyp detection using convolutional neural networks. In: Proceedings of SPIE medical imaging, March 2016, San Diego, CA, USA, vol 9785
Iakovidis DK, Maroulis DE, Karkanis SA (2006) An intelligent system for automatic detection of gastrointestinal adenomas in video endoscopy. Comput Biol Med 36(10):1084–1103
Gokturk SB, Tomasi C, Acar B, Beaulieu CF, Paik DS, Jeffrey RJ, Yee J, Napel S (2001) A statistical 3-D pattern processing method for computer-aided detection of polyps in CT colonography. IEEE Trans Med Imaging 20:1251–1260
Ameling S, Wirth S, Paulus D, Lacey G, Vilarino F (2009) Texture-based polyp detection in colonoscopy. In: Bildver-arbeitung fur die Medizin 2009, Informatik Aktuell. Springer, Berlin, pp 346–350
Park SY, Sargent D, Spofford I, Vosburgh KG, Rahim YA (2012) A colon video analysis framework for polyp detection. IEEE Trans Biomed Eng 59(5):1408–1418
Yi W, Tavanapong W, Wong J, Oh J, de Groen PC (2014) Part-based multiderivative edge cross-sectional profiles for polyp detection in colonoscopy. IEEE J Biomed Health Inform 18(4):1379–1389
Tajbakhsh N, Gurudu SR, Liang J (2016) Automated polyp detection in colonoscopy videos using shape and context information. IEEE Trans Med Imaging 35(2):630–644
Liedlgruber M, Uhl A (2011) Computer-aided decision support systems for endoscopy in the gastrointestinal tract: a review. IEEE Rev Biomed Eng 4:73–88
Hafner M, Brunauer L, Payer H, Resch R, Gangl A, Uhl A, Wrba F, Vecsei A (2010) Computer-aided classification of zoom-endoscopic images using Fourier filters. IEEE Trans Inf Technol Bio-med 14(4):958–970
Karkanis SA, Iakovidis DK, Maroulis DE, Karras DA, Tzivras M (2003) Computer-aided tumor detection in endoscopic video using color wavelet features. IEEE Trans Inf Technol Biomed 7(3):141–152
Vassilis K, Maria B (2007) An adaptive neuro-fuzzy approach for the diagnosis in wireless capsule endoscopy imaging. Int J Inf Technol 13(1):46–56
Alexandre LA, Nobre N, Casteleiro J (2008) Color and position versus texture feature for endoscopic polyp detection. In: Proceedings of the 1st international conference on biomedical engineering and informatics (BMEI’08), Sanya, China, May 2008, vol 2, pp 38–42
Li B, Fan Y, Meng MQH, Qi L (2009) Intestinal polyp recognition in capsule endoscopy images using color and shape features. In: Proceedings of the 2009 IEEE international conference on robotics and biomimetic (ROBIO’09), December 2009, Guilin, China, pp 1490–1494
Tajbakhsh N, Gurudu SR, Liang J (2014) Automatic polyp detection using global geometric constraints and local intensity variation patterns. In: Proceedings of the international conference on medical image computing and computer-assisted intervention (MICCAI’14), Springer International Publishing
Billah M, Waheed S, Rahman MM (2017) An automatic gastrointestinal polyp detection system in video endoscopy using fusion of color wavelet and convolutional neural network features. Int J Biomed Imaging 2017:9545920
Mostafiz R, Hasan M, Hossain I, Rahman MM (2019) An intelligent system for gastrointestinal polyp detection in endoscopic video using a fusion of bidimensional empirical mode decomposition and convolutional neural network features. Int J Imaging Syst Technol 30:224–233
Ribeiro E, Uhl A, Hafner M (2016) Colonic polyp classification with convolutional neural networks. In: Proceedings of the IEEE 29th international symposium on computer-based medical systems (CBMS’16), June 2016, Dublin, Ireland
Ribeiro E, Uhl A, Wimmer G, Hafner M (2016) Exploring deep learning and transfer learning for colonic polyp classification. Comput Math Methods Med 2016:6584725
Bernal J, Tajkbaksh N, Sanchez FJ et al (2017) Comparative validation of polyp detection methods in video colonoscopy: results from the MICCAI 2015 Endoscopic Vision Challenge. IEEE Trans Med Imaging 36(6):1231–1249
Mesejo P, Pizarro D, Abergel A et al (2016) Colonoscopy dataset: computer-aided classification of gastrointestinal lesions in regular colonoscopy. IEEE Trans Med Imaging 35(9):2051–2063
Damerval C, Meignen S, Perrier V (2005) A fast algorithm for bidimensional EMD. IEEE Signal Process Lett 12(10):701–704
Linderhed A (2002) 2D empirical mode decompositions in the spirit of image compression. In: Proceedings of SPIE, Arizona, USA, pp 1–8
Unes J, Bouaoune Y, Delechelle E, Niang O, Bunel P (2003) Image analysis by bidimensional empirical mode decomposition. Image Vis Comput 21(12):1019–1026
Liu Z, Song P, Zhang J, Wang J (2007) Bidimensional empirical mode decomposition for the fusion of multispectral and panchromatic images. Int J Remote Sens 28:4081–4093
Tang X, Li W, Li X, Ma W, Dang X (2020) Motor imagery EEG recognition based on conditional optimization empirical mode decomposition and multi-scale convolutional neural network. Expert Syst Appl 149:113285
Haralick RM, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern 3(6):610–621
Li S, Kwok JT, Zhu H, Wang Y (2003) Texture classification using the support vector machines. Pattern Recognit 36(12):2883–2893
El-Naqa I, Yang Y, Wernick MN, Galatsanos NP, Nishikawa RM (2002) A support vector machine approach for detection of microcalcifications. IEEE Trans Med Imaging 21(12):1552–1563
Graf AB, Wallraven C (2002) Multi-class SVMs for image classification using feature tracking. Technical Report 099. MaxPlanck Institute for Biological Cybernetics. http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.331.4169&rep=rep1&type=pdf
Zhu R, Zhang R, Xue D (2015) Lesion detection of endoscopy images based on convolutional neural network features. In: Proceedings of the 8th international congress on image and signal processing (CISP’15), October 2015, Shenyang, China, pp 372–376
Baxt WG (1995) Application of artificial neural networks to clinical medicine. The Lancet 346(8983):1135–1138
Zou Y, Li L, Wang Y, Yu J, Li Y, Deng WJ (2015) Classifying digestive organs in wireless capsule endoscopy images based on deep convolutional neural network. In: Proceedings of the IEEE international conference on digital signal processing (DSP’15), July 2015, Singapore, pp 1274–1278
Acknowledgements
We are very grateful to Dr. Md. Farhan Matin, Associate Professor, Department of Radiology and Imaging, Uttara Adhunik Medical College and Hospital, Dhaka, Bangladesh for his valuable suggestions.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval (involvement of animals)
This article does not contain any studies with animals performed by any of the authors.
Ethical approval (involvement of human subjects)
This article does not contain any studies with human participants performed by any of the authors. The datasets used in the paper are publicly available. Proper acknowledgments with citation guidelines are maintained for the use of these datasets.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mostafiz, R., Rahman, M.M. & Uddin, M.S. Gastrointestinal polyp classification through empirical mode decomposition and neural features. SN Appl. Sci. 2, 1143 (2020). https://doi.org/10.1007/s42452-020-2944-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42452-020-2944-4