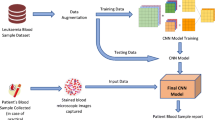
Abstract
Acute Lymphoblastic Leukemia (ALL) is one of the most common types of cancer globally, and the invasive tests used to diagnose it are costly and time-consuming. Peripheral Blood Smear (PBS) images are often preferred for the initial screening of cancer for the diagnosis of ALL. The manual search of PBS images for cancer diagnosis in laboratories is prone to errors due to certain factors such as interoperability errors and human fatigue. Moreover, there are serious problems in using trained data due to the non-specific nature of the signs and symptoms of blood smear images and the complex morphological structure. Advanced techniques have outstripped handcrafted and traditional methods for solving image processing problems. In this paper, an improved ResNet50 Convolutional Neural Network (CNN) model that uses a hybridization of Particle Swarm Optimization (PSO) to detect ALL and its subtypes, is proposed. The last 4 layers have been removed from the ResNet50 backbone model and 11 new layers have been added instead. The model is trained on an augmented dataset by applying color threshold-based segmentation in the Hue/saturation/value (HSV) color space to a publicly available dataset with 3256 PBS images. It has been observed that the performance obtained by classifying the trained improved ResNet50 model outperforms in terms of 99.65% accuracy. It can be said that the proposed method can help to distinguish between different categories of ALL and to determine the diagnosis and treatment protocol for laboratory workers.










Similar content being viewed by others
Data Availability
The dataset used in this paper was obtained from the publicly shared Kaggle platform [11]. The dataset consists of four classes, partially balanced Benign, Early pre-B, Pre-B, and Pro-B. Segmentation and pre-processing steps are applied to highlight the morphological ALL features of the dataset samples and to provide interclass balancing.
References
Johnson, D. T., Zhou, J., Kroll, A. V., Fang, R. H., Yan, M., Xiao, C., Chen, X., Zhang, L., & Zhang, D. E. (2022). Acute myeloid leukemia cell membrane-coated nanoparticles for cancer vaccination immunotherapy. Leukemia, 36(4), 994–1005. https://doi.org/10.1038/s41375-021-01432-w
Kabra, A., & Bushweller, J. (2022). The intrinsically disordered proteins MLLT3 (AF9) and MLLT1 (ENL)–multimodal transcriptional switches with roles in normal hematopoiesis, MLL fusion leukemia, and kidney cancer. Journal of Molecular Biology, 434(1), 167117. https://doi.org/10.1016/j.jmb.2021.167117
Kulkarni, U. P., Selvarajan, S., Lionel, S., Prakash, M. A., Palani, H. K., Balasundaram, N., Arvind Venkataraman, A., Korula, A., Devasia, A. J., Fouzia, N. A., Janet, N. B., Nair, S. C., Abraham, A., Mani, T., Lakshmanan, J., Arunachalam, A. K., Balasubramanian, P., Biju George, B., & Vikram Mathews, V. (2022). Real world data with concurrent retinoic acid and arsenic trioxide for the treatment of acute promyelocytic leukemia. Blood Cancer Journal, 12(1), 22. https://doi.org/10.1182/blood-2021-146362
Dong, Y., Shi, O. M., Zeng, Q. X., Lu, X. Q., Wang, W., Li, Y., & Wang, Q. (2020). Leukemia incidence trends at the global, regional, and national level between 1990 and 2017. Experimental Hematology & Oncology, 9, 1–11. https://doi.org/10.1186/s40164-020-00170-6
Das, P. K., Diya, V. A., Meher, S., Panda, R., & Abraham, A. (2022). A systematic review on recent advancements in deep and machine learning based detection and classification of acute lymphoblastic leukemia. IEEE Access, 10, 81741–81763. https://doi.org/10.1109/ACCESS.2022.3196037
Navya, K. T., Prasad, K., & Singh, B. M. K. (2022). Analysis of red blood cells from peripheral blood smear images for anemia detection: a methodological review. Medical & Biological Engineering & Computing, 60(9), 2445–2462. https://doi.org/10.1007/s11517-022-02614-z
Singh, K., Tiwari, D., Boddu, R., Somasundarum, V., & Mishra, K. (2022). Hematogones: The supreme mimicker and a cytomorphological confounder in acute lymphoblastic leukemia. Journal of Laboratory Physicians, 15(02), 212–216. https://doi.org/10.1055/s-0042-1757586
Özbay, E. (2023). An active deep learning method for diabetic retinopathy detection in segmented fundus images using artificial bee colony algorithm. Artificial Intelligence Review, 56(4), 3291–3318. https://doi.org/10.1007/s10462-022-10231-3
Shahzad, M., Umar, A. I., Shirazi, S. H., Khan, Z., Khan, A., Assam, M., Mohamed, A., & Attia, E. A. (2022). Identification of anemia and its severity level in a peripheral blood smear using 3-tier deep neural network. Applied Sciences, 12(10), 5030. https://doi.org/10.3390/app12105030
Greenberg, P. L. (2023). Myelodysplastic syndromes, thy name is heterogeneity. British Journal of Haematology, 201(3), 381–382. https://doi.org/10.1111/bjh.18649
Gupta, R., Gehlot, S., & Gupta, A. (2022). C-NMC: B-lineage acute lymphoblastic leukaemia: A blood cancer dataset. Medical Engineering & Physics, 103, 103793. https://doi.org/10.1016/j.medengphy.2022.103793
Cengil, E., Çınar, A., & Yıldırım, M. (2022). A hybrid approach for efficient multi-classification of white blood cells based on transfer learning techniques and traditional machine learning methods. Concurrency and Computation: Practice and Experience, 34(6), e6756. https://doi.org/10.1002/cpe.6756
Özbay, E., & Özbay, F. A. (2023). Interpretable pap-smear image retrieval for cervical cancer detection with rotation invariance mask generation deep hashing. Computers in Biology and Medicine, 154, 106574. https://doi.org/10.1016/j.compbiomed.2023.106574
Wang, G. G., Lu, M., Dong, Y. Q., & Zhao, X. J. (2016). Self-adaptive extreme learning machine. Neural Computing and Applications, 27, 291–303. https://doi.org/10.1007/s00521-015-1874-3
Wang, Y., Qiao, X., & Wang, G. G. (2022). Architecture evolution of convolutional neural network using monarch butterfly optimization. Journal of Ambient Intelligence and Humanized Computing. https://doi.org/10.1007/s12652-022-03766-4
Cui, Z., Xue, F., Cai, X., Cao, Y., Wang, G. G., & Chen, J. (2018). Detection of malicious code variants based on deep learning. IEEE Transactions on Industrial Informatics, 14(7), 3187–3196. https://doi.org/10.1109/TII.2018.2822680
Katz, B. Z., Feldman, M. D., Tessema, M., Benisty, D., Toles, G. S., Andre, A., Shtreker, B., Paz, F. M., Edwards, J., Jengehino, D., Bagg, A., Avivi, I., & Pozdnyakova, O. (2021). Evaluation of Scopio labs X100 full field PBS: The first high-resolution full field viewing of peripheral blood specimens combined with artificial intelligence-based morphological analysis. International Journal of Laboratory Hematology, 43(6), 1408–1416. https://doi.org/10.1111/ijlh.13681
Fasihi, M., Nadimi-Shahraki, M. H., & Jannesari, A. (2021). A shallow 1-D convolution neural network for fetal state assessment based on cardiotocogram. SN Computer Science, 2(4), 287. https://doi.org/10.1007/s42979-021-00694-6
Fasihi, M., & Nadimi-Shahraki, M. H. (2020, August). Multi-class cardiovascular diseases diagnosis from electrocardiogram signals using 1-D convolution neural network. In: 2020 IEEE 21st International Conference on Information Reuse and Integration for Data Science (IRI) (pp. 372–378). https://doi.org/10.1109/IRI49571.2020.00060
Alhussan, A. A., Abdelhamid, A. A., Towfek, S. K., Ibrahim, A., Abualigah, L., Khodadadi, N., Khafaga, D. S., Al-Otaibi, S., & Ahmed, A. E. (2023). Classification of breast cancer using transfer learning and advanced al-biruni earth radius optimization. Biomimetics, 8(3), 270. https://doi.org/10.3390/biomimetics8030270
Abualigah, L., Diabat, A., Thanh, C. L., & Khatir, S. (2023). Opposition-based laplacian distribution with prairie dog optimization method for industrial engineering design problems. Computer Methods in Applied Mechanics and Engineering, 414, 116097. https://doi.org/10.1016/j.cma.2023.116097
Abdollahzadeh, B., Gharehchopogh, F. S., & Mirjalili, S. (2021). African vultures optimization algorithm: A new nature-inspired metaheuristic algorithm for global optimization problems. Computers & Industrial Engineering, 158, 107408. https://doi.org/10.1016/j.cie.2021.107408
Ekinci, S., Izci, D., Abualigah, L., & Zitar, R. A. (2023). A modified oppositional chaotic local search strategy based Aquila Optimizer to design an effective controller for vehicle cruise control system. Journal of Bionic Engineering, 20, 1828–1851. https://doi.org/10.1007/s42235-023-00336-y
Abualigah, L., Habash, M., Hanandeh, E. S., Hussein, A. M., Shinwan, M. A., Zitar, R. A., & Jia, H. (2023). Improved reptile search algorithm by salp swarm algorithm for medical image segmentation. Journal of Bionic Engineering, 20, 1766–1790. https://doi.org/10.1007/s42235-023-00332-2
Oyelade, O. N., Ezugwu, A. E. S., Mohamed, T. I., & Abualigah, L. (2022). Ebola optimization search algorithm: A new nature-inspired metaheuristic optimization algorithm. IEEE Access, 10, 16150–16177. https://doi.org/10.1109/ICECET52533.2021.9698813
Abualigah, L., & Diabat, A. (2022). Improved multi-core arithmetic optimization algorithm-based ensemble mutation for multidisciplinary applications. Journal of Intelligent Manufacturing, 34, 1833–1874. https://doi.org/10.1007/s10845-021-01877-x
Shen, B., Khishe, M., & Mirjalili, S. (2023). Evolving marine predators algorithm by dynamic foraging strategy for real-world engineering optimization problems. Engineering Applications of Artificial Intelligence, 123, 106207. https://doi.org/10.1016/j.engappai.2023.106207
Rezaei, F., Safavi, H. R., Abd Elaziz, M., & Mirjalili, S. (2023). GMO: Geometric mean optimizer for solving engineering problems. Soft Computing, 27(15), 10571–10606. https://doi.org/10.1007/s00500-023-08202-z
Mirjalili, S., Mirjalili, S. M., & Lewis, A. (2014). Grey wolf optimizer. Advances in Engineering Software, 69, 46–61. https://doi.org/10.1016/j.advengsoft.2013.12.007
Nadimi-Shahraki, M. H., Taghian, S., & Mirjalili, S. (2021). An improved grey wolf optimizer for solving engineering problems. Expert Systems with Applications, 166, 1–44. https://doi.org/10.1016/j.eswa.2020.113917
Sallam, N. M., Saleh, A. I., Ali, H. A., & Abdelsalam, M. M. (2023). An efficient EGWO algorithm as feature selection for B-ALL diagnoses and its subtypes classification using peripheral blood smear images. Alexandria Engineering Journal, 68, 39–66. https://doi.org/10.3390/app122110760
Boldú, L., Merino, A., Alférez, S., Molina, A., Acevedo, A., & Rodellar, J. (2019). Automatic recognition of different types of acute leukaemia in peripheral blood by image analysis. Journal of Clinical Pathology, 72(11), 755–761. https://doi.org/10.1136/jclinpath-2019-205949
Rehman, A., Abbas, N., Saba, T., Rahman, S. I. U., Mehmood, Z., & Kolivand, H. (2018). Classification of acute lymphoblastic leukemia using deep learning. Microscopy Research and Technique, 81(11), 1310–1317. https://doi.org/10.1002/jemt.23139
Al Mamun, A., Hossen, M. J., Tahabilder, A., Musha, A., Hasnat, R., & Saha, S. K. (2022). Acute lymphoblastic leukemia detection approach from peripheral blood smear using color threshold and morphological techniques. International Journal of Electrical and Computer Engineering, 12(4), 3692. https://doi.org/10.11591/ijece.v12i4.pp3692-3699
Mirmohammadi, P., Ameri, M., & Shalbaf, A. (2021). Recognition of acute lymphoblastic leukemia and lymphocytes cell subtypes in microscopic images using random forest classifier. Physical and Engineering Sciences in Medicine, 44, 433–441. https://doi.org/10.1007/s13246-021-00993-5
Acharya, V., & Kumar, P. (2019). Detection of acute lymphoblastic leukemia using image segmentation and data mining algorithms. Medical & Biological Engineering & Computing, 57, 1783–1811. https://doi.org/10.1007/s11517-019-01984-1
Tusar, M. T. H. K., & Anik, R. K. (2022). Automated detection of acute lymphoblastic leukemia subtypes from microscopic blood smear images using deep neural networks. arXiv preprint https://arXiv.org/2208.08992. https://doi.org/10.48550/arXiv.2208.08992
Özbay, E., & Özbay, F. A. (2023). Interpretable features fusion with precision MRI images deep hashing for brain tumor detection. Computer Methods and Programs in Biomedicine, 231, 107387. https://doi.org/10.1016/j.cmpb.2023.107387
Hu, N., Zhang, D., Xie, K., Liang, W., & Hsieh, M. Y. (2022). Graph learning-based spatial-temporal graph convolutional neural networks for traffic forecasting. Connection Science, 34(1), 429–448. https://doi.org/10.1080/09540091.2021.2006607
Dubey, S. R., Singh, S. K., & Chaudhuri, B. B. (2022). Activation functions in deep learning: A comprehensive survey and benchmark. Neurocomputing, 503, 92–108. https://doi.org/10.1016/j.neucom.2022.06.111
Yuan, X., Shen, X., Mehta, S., Li, T., Ge, S., & Zha, Z. (2022). Structure injected weight normalization for training deep networks. Multimedia Systems, 28, 433–444. https://doi.org/10.1007/s00530-021-00793-7
Zhang, X., Li, C., Xu, B., Pan, Z., & Yu, T. (2022). Dropout deep neural network assisted transfer learning for bi-objective Pareto AGC dispatch. IEEE Transactions on Power Systems, 38(2), 1432–1444. https://doi.org/10.1109/TPWRS.2022.3179372
Citak-Er, F., Firat, Z., Kovanlikaya, I., Ture, U., & Ozturk-Isik, E. (2018). Machine-learning in grading of gliomas based on multi-parametric magnetic resonance imaging at 3T. Computers in Biology and Medicine, 99, 154–160. https://doi.org/10.1016/j.compbiomed.2018.06.009
Varedi-Koulaei, S. M., Mohammadi, M., Mohammadi, M. A. M., & Bamdad, M. (2023). Optimal synthesis of the stephenson-ii linkage for finger exoskeleton using swarm-based optimization algorithms. Journal of Bionic Engineering, 20, 1569–1584. https://doi.org/10.1007/s42235-022-00327-5
Wang, D., Tan, D., & Liu, L. (2018). Particle swarm optimization algorithm: An overview. Soft Computing, 22, 387–408. https://doi.org/10.1007/s00500-016-2474-6
Yin, S. H., Luo, Q. F., & Zhou, Y. Q. (2023). IBMSMA: An indicator-based multi-swarm slime mould algorithm for multi-objective truss optimization problems. Journal of Bionic Engineering, 20, 1333–1360. https://doi.org/10.1007/s42235-022-00307-9
Ghaderzadeh, M., Aria, M., Hosseini, A., Asadi, F., Bashash, D., & Abolghasemi, H. (2022). A fast and efficient CNN model for B-ALL diagnosis and its subtypes classification using peripheral blood smear images. International Journal of Intelligent Systems, 37(8), 5113–5133. https://doi.org/10.1002/int.22753
Balasubramanian, K., & Ananthamoorthy, N. P. (2021). Improved adaptive neuro-fuzzy inference system based on modified glowworm swarm and differential evolution optimization algorithm for medical diagnosis. Neural Computing and Applications, 33, 7649–7660. https://doi.org/10.1007/s00521-020-05507-0
Jiang, M., Cheng, L., Qin, F., Du, L., & Zhang, M. (2018). White blood cells classification with deep convolutional neural networks. International Journal of Pattern Recognition and Artificial Intelligence, 32(09), 1857006. https://doi.org/10.1142/S0218001418570069
Şengür, A., Akbulut, Y., Budak, Ü., & Cömert, Z. (2019, September). White blood cell classification based on shape and deep features. In: 2019 International Artificial Intelligence and Data Processing Symposium (IDAP) (pp. 1–4). https://doi.org/10.1109/IDAP.2019.8875945
Liang, G., Hong, H., Xie, W., & Zheng, L. (2018). Combining convolutional neural network with recursive neural network for blood cell image classification. IEEE Access, 6, 36188–36197. https://doi.org/10.1109/ACCESS.2018.2846685
Throngnumchai, K., Lomvisai, P., Tantasirin, C., & Phasukkit, P. (2019, November). Classification of white blood cell using deep convolutional neural network. In: 2019 12th Biomedical Engineering International Conference. (pp. 1–4). IEEE. https://doi.org/10.1109/BMEiCON47515.2019.8990301
Habibzadeh, M., Jannesari, M., Rezaei, Z., Baharvand, H., & Totonchi, M. (2018, April). Automatic white blood cell classification using pre-trained deep learning models: Resnet and inception. In: Tenth International Conference On Machine Vision, Vienna, Austria, (10696),(pp. 274–281). SPIE. https://doi.org/10.1117/12.2311282
Patil, A. M., Patil, M. D., & Birajdar, G. K. (2021). White blood cells image classification using deep learning with canonical correlation analysis. IRBM, 42(5), 378–389. https://doi.org/10.1016/j.irbm.2020.08.005
Özyurt, F. (2020). A fused CNN model for wbc detection with mrmr feature selection and extreme learning machine. Soft Computing, 24(11), 8163–8172. https://doi.org/10.1007/s00500-019-04383-8
Toğaçar, M., Ergen, B., & Cömert, Z. (2020). Classification of white blood cells using deep features obtained from convolutional neural network models based on the combination of feature selection methods. Applied Soft Computing, 97(B), 106810. https://doi.org/10.1016/j.asoc.2020.106810
Çınar, A., & Tuncer, S. A. (2021). Classification of lymphocytes, monocytes, eosinophils, and neutrophils on white blood cells using hybrid Alexnet-GoogleNet-SVM. SN Applied Sciences, 3, 1–11. https://doi.org/10.1007/s42452-021-04485-9
Ridoy, M. A. R., & Islam, M. R. (2020, November). An automated approach to white blood cell classification using a lightweight convolutional neural network. In: 2020 2nd International Conference on Advanced Information and Communication Technology (ICAICT), Dhaka, Bangladesh (pp. 480–483). IEEE. https://doi.org/10.1109/ICAICT51780.2020.9333512
Edraki, A., & Razminia, A. (2018). Classification of white blood cells using convolutional neural network. ISMJ, 21(1), 65–80. https://doi.org/10.29252/ismj.21.1.65
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
No human or animal studies were conducted by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Özbay, E., Özbay, F.A. & Gharehchopogh, F.S. Peripheral Blood Smear Images Classification for Acute Lymphoblastic Leukemia Diagnosis with an Improved Convolutional Neural Network. J Bionic Eng (2023). https://doi.org/10.1007/s42235-023-00441-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s42235-023-00441-y