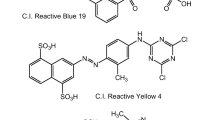
Abstract
Dyes color various materials, including leather, paper, cosmetics, and textiles. Wastewater contains different dyes that have negative impacts on humans and the environment. Dye-containing wastewater has been treated using several well-known procedures, including physicochemical and biological degradation. Interestingly, the breakdown of industrial dyes by fungus, yeast, and algae is considered an innovative treatment technology to control chemicals and secondary water pollution. The present review discusses the degradation of industrial dyes using a microbial community with a special focus on the microbial degradation mechanism of dye molecules. The influences of different parameters (e.g., pH, temperature, concentrations of dyes, soluble salts, the effects of CO2 and nitrogen, and dye structure) have been discussed to achieve optimal dye degradation potential of microbes. In addition, the role of genetically modified organisms (GMOs) and the utilization of various enzymes in dyes biodegradation are also highlighted along with prospects. This review also critically sketches the most recent advancements and advanced bioreactors for dye degradation. It is anticipated that the treatment of industrial dyes using microbial community is an advanced technology to establish a pollution-free natural environment and successfully achieve one of the sustainable development goals (SDGs).






Similar content being viewed by others
Data Availability
All relevant data have been mentioned in the form of Tables.
References
Holkar CR, Jadhav AJ, Pinjari DV, Mahamuni NM, Pandit AB (2016) A critical review on textile wastewater treatments: possible approaches. J Environ Manag 182:351–366. https://doi.org/10.1016/j.jenvman.2016.07.090
Rani B, Maheshwari R, Yadav R, Pareek D, Sharma A (2013) Resolution to provide safe drinking water for sustainability of future perspectives. Res J Chem Env Sci 1:50–54
Chen HL, Burns LD (2006) Environmental analysis of textile products. Cloth Text Res J 24:248–261. https://doi.org/10.1177/0887302X06293065
Bharathi KS, Ramesh ST (2013) Removal of dyes using agricultural waste as low-cost adsorbents: a review. Appl Water Sci 3:773–790. https://doi.org/10.1007/s13201-013-0117-y
Olukanni OD, Osuntoki AA, Gbenle GO (2006) Textile effluent biodegradation potentials of textile effluent-adapted and non-adapted bacteria, African. J Biotechnol 5:1980–1984. https://doi.org/10.4314/ajb.v5i20.55924
Çelik L (2012) Biodegradation of reactive red 195 azo dye by the bacterium Rhodopseudomonas palustris 51ATA, African. J Microbiol Res 6:120–126. https://doi.org/10.5897/ajmr11.1059
Park C, Lee M, Lee B, Kim SW, Chase HA, Lee J, Kim S (2007) Biodegradation and biosorption for decolorization of synthetic dyes by Funalia trogii. Biochem Eng J 36:59–65. https://doi.org/10.1016/j.bej.2006.06.007
Mohan SV, Bhaskar YV, Karthikeyan J (2004) Biological decolourization of simulated azo dye in aqueous phase by algae Spirogyra species. Int J Environ Pollut 21:211–222. https://doi.org/10.1504/IJEP.2004.004190
Kuhad RC, Sood N, Tripathi KK, Singh A, Ward OP (2004) Developments in microbial methods for the treatment of dye effluents. Adv Appl Microbiol 56:185–213. https://doi.org/10.1016/S0065-2164(04)56006-9
Xu M, Guo J, Sun G (2007) Biodegradation of textile azo dye by Shewanella decolorationis S12 under microaerophilic conditions. Appl Microbiol Biotechnol 76:719–726. https://doi.org/10.1007/s00253-007-1032-7
Cao XL, Yan YN, Zhou FY, Sun SP (2020) Tailoring nanofiltration membranes for effective removing dye intermediates in complex dye-wastewater. J Membr Sci 595. https://doi.org/10.1016/j.memsci.2019.117476
Varjani S, Upasani VN (2019) Comparing bioremediation approaches for agricultural soil affected with petroleum crude: a case study. Indian J Microbiol 59:356–364. https://doi.org/10.1007/s12088-019-00814-0
Swaminathan K, Sandhya S, Sophia AC, Pachhade K, Subrahmanyam YV (2003) Decolorization and degradation of H-acid and other dyes using ferrous – hydrogen peroxide system. Chemosphere 50:619–625
Behnajady MA (2004) Photodestruction of Acid Orange 7 ( AO7 ) in aqueous solutions by UV / H 2 O 2 : influence of operational parameters. Chemosphere 55:129–134. https://doi.org/10.1016/j.chemosphere.2003.10.054
Arslan I, Akmehmet I, Bahnemann DW (2002) Advanced oxidation of a reactive dyebath effluent : comparison of O 3 , H 2 O 2 / UV-C and TiO 2 / UV-A processes. Water res 36:1143–1154
Al-kdasi A, Idris A (2005) Treatment of textile wastewater by advanced oxidation processes – a review. Global nest: the Int J 6:222–230
Stolz A (2001) Basic and applied aspects in the microbial degradation of azo dyes. Appl Microbiol Biotechnol 56:69–80. https://doi.org/10.1007/s002530100686
McMullan G, Meehan C, Conneely A, Kirby N, Robinson T, Nigam P, Banat IM, Marchant R, Smyth WF (2001) Microbial decolourization and degradation of textile dyes. Appl Microbiol Biotechnol 56:81–87. https://doi.org/10.1007/s002530000587
Banerjee UC P. Education, critical reviews in environmental science and technology(2007) Removal of dyes from the effluent of textile and dyestuff manufacturing industry: a review of emerging techniques with reference to biological treatment. Crit Rev Environ Sci Technol:219–238. https://doi.org/10.1080/10643380590917932
Van Der Zee FP, Villaverde S (2005) Combined anaerobic – aerobic treatment of azo dyes — a short review of bioreactor studies. Water res 39:1425–1440. https://doi.org/10.1016/j.watres.2005.03.007
Banat IM, Nigam P, Singh D, Marchant R (1997) Microbial decolorization of textile-dye- containing effluents : a review. Bioresour Technol 58:217–227
Imran M, Crowley DE, Khalid A, Hussain S, Mumtaz MW, Arshad M (2015) Microbial biotechnology for decolorization of textile wastewaters. Rev Environ Sci Biotechnol 14:73–92. https://doi.org/10.1007/s11157-014-9344-4
Shindhal T, Rakholiya P, Varjani S, Pandey A, Ngo HH, Guo W, Ng HY, Taherzadeh MJ (2021) A critical review on advances in the practices and perspectives for the treatment of dye industry wastewater. Bioengineered 12:70–87. https://doi.org/10.1080/21655979.2020.1863034
Varjani SJ, Upasani VN (2017) A new look on factors affecting microbial degradation of petroleum hydrocarbon pollutants. Int Biodeterior Biodegrad 120:71–83. https://doi.org/10.1016/j.ibiod.2017.02.006
Kumar A, Kumar A, Singh R, Singh R, Pandey S, Rai A, Singh VK, Rahul B (2019) Genetically engineered bacteria for the degradation of dye and other organic compounds. In: Abatement of environmental pollutants. Elsevier, pp 331–350. https://doi.org/10.1016/B978-0-12-818095-2.00016-3
Jin RF, Zhou JT, Zhang AL, Wang J (2008) Bioaugmentation of the decolorization rate of acid red GR by genetically engineered microorganism Escherichia coli JM109 (pGEX-AZR). World J Microbiol Biotechnol 24:23–29. https://doi.org/10.1007/s11274-007-9433-4
Al-Amrani WA, Lim PE, Seng CE, Wan Ngah WS (2014) Factors affecting bio-decolorization of azo dyes and COD removal in anoxic-aerobic REACT operated sequencing batch reactor. J Taiwan Inst Chem Eng 45:609–616. https://doi.org/10.1016/j.jtice.2013.06.032
Khan R, Bhawana P, Fulekar MH (2013) Microbial decolorization and degradation of synthetic dyes: a review. Rev Environ Sci Biotechnol 12:75–97. https://doi.org/10.1007/s11157-012-9287-6
Varjani S, Rakholiya P, Ng HY, You S, Teixeira JA (2020) Microbial degradation of dyes: an overview. Bioresour Technol 314. https://doi.org/10.1016/j.biortech.2020.123728
Ajaz M, Shakeel S, Rehman A (2020) Microbial use for azo dye degradation—a strategy for dye bioremediation. Int Microbiol 23:149–159. https://doi.org/10.1007/s10123-019-00103-2
Ihsanullah I, Jamal A, Ilyas M, Zubair M, Khan G, Atieh MA (2020) Bioremediation of dyes: current status and prospects. J Water Process Eng 38:101680. https://doi.org/10.1016/j.jwpe.2020.101680
R. Shyamala Gowri, R. Vijayaraghavan, P. Meenambigai, Microbial degradation of reactive dyes-a review, 2014. http://www.ijcmas.com.
Pavlikova M (2020) Microbial degradation of wool industry wastewater. IOP Conf Ser Earth Environ Sci 444. https://doi.org/10.1088/1755-1315/444/1/012042
Gürses A, Güneş K, Şahin E (2020) Removal of dyes and pigments from industrial effluents. In: Green chemistry and water remediation: Research and applications. Elsevier, pp 135–187. https://doi.org/10.1016/B978-0-12-817742-6.00005-0
Wang H, Su JQ, Zheng XW, Tian Y, Xiong XJ, Zheng TL (2009) Bacterial decolorization and degradation of the reactive dye Reactive Red 180 by Citrobacter sp. CK3. Int Biodeterior Biodegrad 63:395–399. https://doi.org/10.1016/j.ibiod.2008.11.006
Chung YC, Chen CY (2009) Degradation of azo dye reactive violet 5 by TiO2 photocatalysis. Environ Chem Lett 7:347–352. https://doi.org/10.1007/s10311-008-0178-6
Manzoor J, Sharma M (2019) Impact of textile dyes on human health and environment. IGI Global, pp 162–169. https://doi.org/10.4018/978-1-7998-0311-9.ch008
Ben Mansour H, Ayed-Ajmi Y, Mosrati R, Corroler D, Ghedira K, Barillier D, Chekir-Ghedira L (2010) Acid violet 7 and its biodegradation products induce chromosome aberrations, lipid peroxidation, and cholinesterase inhibition in mouse bone marrow. Environ Sci Pollut Res 17:1371–1378. https://doi.org/10.1007/s11356-010-0323-1
Fernandes FH, de Aragão Umbuzeiro G, Salvadori DMF (2019) Genotoxicity of textile dye CI Disperse Blue 291 in mouse bone marrow. Mutat Res Genet Toxicol Environ Mutagen 837:48–51. https://doi.org/10.1016/j.mrgentox.2018.10.003
Ferraz ERA, Oliveira GAR, Grando MD, Lizier TM, Zanoni MVB, Oliveira DP (2013) Photoelectrocatalysis based on Ti/TiO2 nanotubes removes toxic properties of the azo dyes Disperse Red 1, Disperse Red 13 and Disperse Orange 1 from aqueous chloride samples. J Environ Manag 124:108–114. https://doi.org/10.1016/j.jenvman.2013.03.033
Singh R, Gautam N, Mishra A, Gupta R (2011) Heavy metals and living systems: an overview. Indian J Pharmacol 43:246–253. https://doi.org/10.4103/0253-7613.81505
Karimifard S, Alavi Moghaddam MR (2018) Application of response surface methodology in physicochemical removal of dyes from wastewater: a critical review. Sci Total Environ 640–641:772–797. https://doi.org/10.1016/j.scitotenv.2018.05.355
Ayed L, El Ksibi I, Charef A, El Mzoughi R (2021) Hybrid coagulation-flocculation and anaerobic-aerobic biological treatment for industrial textile wastewater: pilot case study. J Text Inst 112:200–206. https://doi.org/10.1080/00405000.2020.1731273
Jafari Chermahini Z, Najafi Chermahini A, Dabbagh HA, Rezaei B, Irannejad N (2017) Synthesis of new dyes containing double tetrazole groups for sensitization of TiO2 nanoparticles in dye-sensitized solar cells. J Iran Chem Soc 14:1549–1556. https://doi.org/10.1007/s13738-017-1096-y
Donkadokula NY, Kola AK, Naz I, Saroj D (2020) A review on advanced physico-chemical and biological textile dye wastewater treatment techniques. Rev Environ Sci Biotechnol 19:543–560. https://doi.org/10.1007/s11157-020-09543-z
Kumar P, Agnihotri R, Wasewar KL, Uslu H, Yoo CK (2012) Status of adsorptive removal of dye from textile industry effluent. Desalin Water Treat 50:226–244. https://doi.org/10.1080/19443994.2012.719472
Varjani SJ, Rana DP, Jain AK, Bateja S, Upasani VN (2015) Synergistic ex-situ biodegradation of crude oil by halotolerant bacterial consortium of indigenous strains isolated from on shore sites of Gujarat, India. Int Biodeterior Biodegrad 103:116–124. https://doi.org/10.1016/j.ibiod.2015.03.030
Bhatia D, Sharma NR, Singh J, Kanwar RS (2017) Biological methods for textile dye removal from wastewater: a review. Crit Rev Environ Sci Technol 47:1836–1876. https://doi.org/10.1080/10643389.2017.1393263
Varjani S, Upasani VN (2019) Influence of abiotic factors, natural attenuation, bioaugmentation and nutrient supplementation on bioremediation of petroleum crude contaminated agricultural soil. J Environ Manag 245:358–366. https://doi.org/10.1016/j.jenvman.2019.05.070
Ali H (2010) Biodegradation of synthetic dyes - a review, water. Air Soil Pollut 213:251–273. https://doi.org/10.1007/s11270-010-0382-4
Vijaykumar MH, Vaishampayan PA, Shouche YS, Karegoudar TB (2007) Decolourization of naphthalene-containing sulfonated azo dyes by Kerstersia sp. strain VKY1. Enzym Microb Technol 40:204–211. https://doi.org/10.1016/j.enzmictec.2006.04.001
Lin YH, Leu JY (2008) Kinetics of reactive azo-dye decolorization by Pseudomonas luteola in a biological activated carbon process. Biochem Eng J 39:457–467. https://doi.org/10.1016/j.bej.2007.10.015
Coughlin MF, Kinkle BK, Bishop PL (2002) Degradation of acid orange 7 in an aerobic biofilm. Chemosphere 46:11–19. https://doi.org/10.1016/S0045-6535(01)00096-0
Kishor R, Purchase D, Saratale GD, Ferreira LFR, Bilal M, Iqbal HMN, Bharagava RN (2021) Environment friendly degradation and detoxification of Congo red dye and textile industry wastewater by a newly isolated Bacillus cohnni (RKS9). Environ Technol Innov 22:101425. https://doi.org/10.1016/j.eti.2021.101425
Pereira LDO, Lelo RV, Coelho GC, Magalhães F (2019) Degradation of textile dyes from synthetic and wastewater samples using TiO2/C/Fe magnetic photocatalyst and TiO2. J Iran Chem Soc 16:2281–2289. https://doi.org/10.1007/s13738-019-01694-3
Chen KC, Wu JY, Liou DJ, Hwang SCJ (2003) Decolorization of the textile dyes by newly isolated bacterial strains. J Biotechnol 101:57–68. https://doi.org/10.1016/S0168-1656(02)00303-6
Pajerski W, Ochonska D, Brzychczy-wloch M, Indyka P (2019) Attachment efficiency of gold nanoparticles by Gram-positive and Gram-negative bacterial strains governed by surface charges. J Nanopart Res 21:1–12
Ezhilarasu A (2016) International Journal of Advanced Research in Biological Sciences Textile industry Dye degrading by bacterial strain Bacillus sp. Int J Adv Res Biol Sci 3:211–226
Kishor R, Purchase D, Saratale GD, Romanholo Ferreira LF, Hussain CM, Mulla SI, Bharagava RN (2021) Degradation mechanism and toxicity reduction of methyl orange dye by a newly isolated bacterium Pseudomonas aeruginosa MZ520730. J Water Process Eng 43:102300. https://doi.org/10.1016/j.jwpe.2021.102300
Yu Z, Wen X (2005) Screening and identification of yeasts for decolorizing synthetic dyes in industrial wastewater. Int Biodeterior Biodegrad 56:109–114. https://doi.org/10.1016/j.ibiod.2005.05.006
Jadhav UU, Dawkar VV, Ghodake GS, Govindwar SP (2008) Biodegradation of Direct Red 5B, a textile dye by newly isolated Comamonas sp. UVS. J Hazard Mater 158:507–516. https://doi.org/10.1016/j.jhazmat.2008.01.099
Bellut K, Krogerus K, Arendt EK (2020) Lachancea fermentati strains isolated from kombucha : fundamental insights , and practical application in low alcohol beer brewing. Front Microbiol 11:1–21. https://doi.org/10.3389/fmicb.2020.00764
Meehan C, Banat IM, McMullan G, Nigam P, Smyth F, Marchant R (2000) Decolorization of Remazol Black-B using a thermotolerant yeast, Kluyveromyces marxianus IMB3. Environ Int 26:75–79. https://doi.org/10.1016/S0160-4120(00)00084-2
Saratale GD, Der Chen S, Lo YC, Saratale RG, Chang JS (2008) Outlook of biohydrogen production from lignocellulosic feedstock using dark fermentation - a review. J Sci Ind Res (India) 67:962–979
Safarikova M, Ptáčková L, Kibriková I, Šafařík I (2005) Biosorption of water-soluble dyes on magnetically modified Saccharomyces cerevisiae subsp. uvarum cells. Chemosphere 59:831–835. https://doi.org/10.1016/j.chemosphere.2004.10.062
Kiayi Z, Lotfabad TB, Heidarinasab A, Shahcheraghi F (2019) Microbial degradation of azo dye carmoisine in aqueous medium using Saccharomyces cerevisiae ATCC 9763. J Hazard Mater 373:608–619. https://doi.org/10.1016/j.jhazmat.2019.03.111
Rafii F, Franklin W, Cerniglia CE (1990) Azoreductase activity of anaerobic bacteria isolated from human intestinal microflora. Appl Environ Microbiol 56:2146–2151. https://doi.org/10.1128/aem.56.7.2146-2151.1990
Rodríguez Couto S (2009) Dye removal by immobilized fungi. Biotechnol Adv 27:227–235. https://doi.org/10.1016/j.biotechadv.2008.12.001
Li S, Huang J, Chen G, Parkin IP (2019) Nanoscale advances remediation by Aspergillus niger biosorbent †:168–176. https://doi.org/10.1039/c8na00132d
Nozaki K, Beh CH, Mizuno M, Isobe T, Shiroishi M, Kanda T, Amano Y (2008) Screening and investigation of dye decolorization activities of basidiomycetes. J Biosci Bioeng 105:69–72. https://doi.org/10.1263/jbb.105.69
Enayatzamir K, Tabandeh F, Yakhchali B, Alikhani HA, Rodríguez Couto S (2009) Assessment of the joint effect of laccase and cellobiose dehydrogenase on the decolouration of different synthetic dyes. J Hazard Mater 169:176–181. https://doi.org/10.1016/j.jhazmat.2009.03.088
Kaushik P, Malik A (2009) Fungal dye decolourization: recent advances and future potential. Environ Int 35:127–141. https://doi.org/10.1016/j.envint.2008.05.010
Wesenberg D, Kyriakides I, Agathos SN (2003) White-rot fungi and their enzymes for the treatment of industrial dye effluents. Biotechnol Adv 22:161–187. https://doi.org/10.1016/j.biotechadv.2003.08.011
Novotný Č, Svobodová K, Kasinath A, Erbanová P (2004) Biodegradation of synthetic dyes by Irpex lacteus under various growth conditions. Int Biodeterior Biodegrad 54:215–223. https://doi.org/10.1016/j.ibiod.2004.06.003
Forss J, Welander U (2009) Decolourization of reactive azo dyes with microorganisms growing on soft wood chips. Int Biodeterior Biodegrad 63:752–758. https://doi.org/10.1016/j.ibiod.2009.05.005
Matavulj M, Molitoris H (2009) Marine fungi: degraders of poly-3-hydroxyalkanoate based plastic materials, Zb. Matice Srp Za Prir Nauk:253–265. https://doi.org/10.2298/zmspn0916253m
El-Rahim WMA, Moawad H, Azeiz AZA, Sadowsky MJ (2021) Biodegradation of azo dyes by bacterial or fungal consortium and identification of the biodegradation products. Egypt J Aquat Res 47:269–276. https://doi.org/10.1016/j.ejar.2021.06.002
Daneshvar N, Ayazloo M, Khataee AR, Pourhassan M (2007) Biological decolorization of dye solution containing malachite green by microalgae Cosmarium sp. Bioresour Technol 98:1176–1182. https://doi.org/10.1016/j.biortech.2006.05.025
Vijayaraghavan K, Yun YS (2007) Utilization of fermentation waste (Corynebacterium glutamicum) for biosorption of Reactive Black 5 from aqueous solution. J Hazard Mater 141:45–52. https://doi.org/10.1016/j.jhazmat.2006.06.081
Omar HH (2008) Algal decolorization and degradation of monoazo and diazo dyes, Pakistan. J Biol Sci 11:1310–1316. https://doi.org/10.3923/pjbs.2008.1310.1316
Khataee AR, Dehghan G (2011) Optimization of biological treatment of a dye solution by macroalgae Cladophora sp. using response surface methodology. J Taiwan Inst Chem Eng 42:26–33. https://doi.org/10.1016/j.jtice.2010.03.007
Alam MA, Wan C, Guo SL, Zhao XQ, Huang ZY, Yang YL, Chang JS, Bai FW (2014) Characterization of the flocculating agent from the spontaneously flocculating microalga Chlorella vulgaris JSC-7. J Biosci Bioeng 118:29–33. https://doi.org/10.1016/j.jbiosc.2013.12.021
Acuner E, Dilek FB (2004) Treatment of tectilon yellow 2G by Chlorella vulgaris. Process Biochem 39:623–631. https://doi.org/10.1016/S0032-9592(03)00138-9
Rodrigues de Almeida EJ, Christofoletti Mazzeo DE, Deroldo Sommaggio LR, Marin-Morales MA, Rodrigues de Andrade A, Corso CR (2019) Azo dyes degradation and mutagenicity evaluation with a combination of microbiological and oxidative discoloration treatments. Ecotoxicol Environ Saf 183:109484. https://doi.org/10.1016/j.ecoenv.2019.109484
Dahlia MEM (2013) Evaluation of non-viable biomass of Laurencia papillosa for decolorization of dye waste water, African. J Biotechnol 12:2215–2223. https://doi.org/10.5897/ajb2013.11975
Ren S, Guo J, Zeng G, Sun G (2006) Decolorization of triphenylmethane, azo, and anthraquinone dyes by a newly isolated Aeromonas hydrophila strain. Appl Microbiol Biotechnol 72:1316–1321. https://doi.org/10.1007/s00253-006-0418-2
Wang H, Zheng XW, Su JQ, Tian Y, Xiong XJ, Zheng TL (2009) Biological decolorization of the reactive dyes Reactive Black 5 by a novel isolated bacterial strain Enterobacter sp. EC3. J Hazard Mater 171:654–659. https://doi.org/10.1016/j.jhazmat.2009.06.050
Dawkar VV, Jadhav UU, Ghodake GS, Govindwar SP (2009) Effect of inducers on the decolorization and biodegradation of textile azo dye Navy blue 2GL by Bacillus sp. VUS, Biodegradation 20:777–787. https://doi.org/10.1007/s10532-009-9266-y
Jirasripongpun K, Nasanit R, Niruntasook J, Chotikasatian B (2007) Decolorization and degradation of CI Reactive Red 195 by Enterobacter sp, Thammasat Int. J Sci Technol 12:6–11
Saratale RG, Saratale GD, Chang JS, Govindwar SP (2009) Decolorization and biodegradation of textile dye Navy blue HER by Trichosporon beigelii NCIM-3326. J Hazard Mater 166:1421–1428. https://doi.org/10.1016/j.jhazmat.2008.12.068
Annuar MSM, Adnan S, Vikineswary S, Chisti Y (2009) Kinetics and energetics of azo dye decolorization by Pycnoporus sanguineus. Water Air Soil Pollut 202:179–188. https://doi.org/10.1007/s11270-008-9968-5
Sarnthima R, Khammuang S, Svasti J (2009) Extracellular ligninolytic enzymes by Lentinus polychrous Lév. under solid-state fermentation of potential agro-industrial wastes and their effectiveness in decolorization of synthetic dyes. Biotechnol Bioprocess Eng 14:513–522. https://doi.org/10.1007/s12257-008-0262-6
Yang XQ, Zhao XX, Liu CY, Zheng Y, Qian SJ (2009) Decolorization of azo, triphenylmethane and anthraquinone dyes by a newly isolated Trametes sp. SQ01 and its laccase. Process Biochem 44:1185–1189. https://doi.org/10.1016/j.procbio.2009.06.015
Ali H, Ahmad W, Haq T (2009) Decolorization and degradation of malachite green by Aspergillus flavus and Alternaria solani, African. J Biotechnol 8:1574–1576. https://doi.org/10.4314/ajb.v8i8.60239
Parikh A, Madamwar D (2005) Textile dye decolorization using cyanobacteria. Biotechnol Lett 27:323–326. https://doi.org/10.1007/s10529-005-0691-7
Do MH, Ngo HH, Guo W, Chang SW, Nguyen DD, Liu Y, Varjani S, Kumar M (2020) Microbial fuel cell-based biosensor for online monitoring wastewater quality: a critical review. Sci Total Environ 712:135612. https://doi.org/10.1016/j.scitotenv.2019.135612
Mishra B, Varjani S, Kumar G, Awasthi MK, Awasthi SK, Sindhu R, Binod P, Rene ER, Zhang Z (2021) Microbial approaches for remediation of pollutants: innovations, future outlook, and challenges. Energy Environ 32:1029–1058. https://doi.org/10.1177/0958305X19896781
Karmakar M (2020) Recent trends in waste water treatment and water resource management. Springer, Singapore. https://doi.org/10.1007/978-981-15-0706-9
Kisand V, Tuvikene L, Nõges T (2001) Role of phosphorus and nitrogen for bacteria and phytopankton development in a large shallow lake. Hydrobiologia 457:187–197. https://doi.org/10.1023/A:1012291820177
Khan I, Saeed K, Ali N, Khan I, Zhang B, Sadiq M (2020) Heterogeneous photodegradation of industrial dyes: an insight to different mechanisms and rate affecting parameters. J Environ Chem Eng 8. https://doi.org/10.1016/j.jece.2020.104364
Li N, Chen G, Zhao J, Yan B, Cheng Z, Meng L, Chen V (2019) Self-cleaning PDA/ZIF-67@PP membrane for dye wastewater remediation with peroxymonosulfate and visible light activation. J Membr Sci 591:117341. https://doi.org/10.1016/j.memsci.2019.117341
Liu N, Xie X, Yang B, Zhang Q, Yu C, Zheng X, Xu L, Li R, Liu J (2017) Performance and microbial community structures of hydrolysis acidification process treating azo and anthraquinone dyes in different stages. Environ Sci Pollut Res 24:252–263. https://doi.org/10.1007/s11356-016-7705-y
Varjani SJ, Upasani VN (2017) Critical review on biosurfactant analysis, purification and characterization using rhamnolipid as a model biosurfactant. Bioresour Technol 232:389–397. https://doi.org/10.1016/j.biortech.2017.02.047
Delpla I, Jung AV, Baures E, Clement M, Thomas O (2009) Impacts of climate change on surface water quality in relation to drinking water production. Environ Int 35:1225–1233. https://doi.org/10.1016/j.envint.2009.07.001
Das A, Mishra S (2017) Removal of textile dye reactive green-19 using bacterial consortium: process optimization using response surface methodology and kinetics study. J Environ Chem Eng 5:612–627. https://doi.org/10.1016/j.jece.2016.10.005
Gianfreda L, Rao MA (2004) Potential of extra cellular enzymes in remediation of polluted soils: a review. Enzym Microb Technol 35:339–354. https://doi.org/10.1016/j.enzmictec.2004.05.006
Kirby N, Marchant R, McMullan G (2000) Decolourisation of synthetic textile dyes by Phlebia tremellosa. FEMS Microbiol Lett 188:93–96. https://doi.org/10.1016/S0378-1097(00)00216-0
Bento RMF, Almeida MR, Bharmoria P, Freire MG, Tavares APM (2020) Improvements in the enzymatic degradation of textile dyes using ionic-liquid-based surfactants. Sep Purif Technol 235:116191. https://doi.org/10.1016/j.seppur.2019.116191
Iark D, dos Reis Buzzo AJ, Garcia JAA, Côrrea VG, Helm CV, Corrêa RCG, Peralta RA, Moreira RDFPM, Bracht A, Peralta RM (2019) Enzymatic degradation and detoxification of azo dye Congo red by a new laccase from Oudemansiella canarii. Bioresour Technol 289:121655. https://doi.org/10.1016/j.biortech.2019.121655
Zille A, Gornacka B, Rehorek A, Cavaco-Paulo A (2005) Degradation of azo dyes by Trametes villosa laccase over long periods of oxidative conditions. Appl Environ Microbiol 71:6711–6718. https://doi.org/10.1128/AEM.71.11.6711-6718.2005
Abadulla E, Tzanov T, Costa S, Cavaco A, Gübitz G (2013) Decolorization and Detoxification of Textile Dyes with a Laccase from Trametes hirsuta. Appl Environ Microbiol 66(2000):3357–3362. https://doi.org/10.1128/aem.66.8.3357-3362.2000
Chang JS, Lin YC (2000) Fed-batch bioreactor strategies for microbial decolorization of azo dye using a Pseudomonas luteola strain. Biotechnol Prog 16:979–985. https://doi.org/10.1021/bp000116z
Maier J, Kandelbauer A, Erlacher A, Cavaco-Paulo A, Gübitz GM (2004) A new alkali-thermostable azoreductase from Bacillus sp. strain SF. Appl Environ Microbiol 70:837–844. https://doi.org/10.1128/AEM.70.2.837-844.2004
Ramalho PA, Scholze H, Cardoso MH, Ramalho MT, Oliveira-Campos AM (2002) Improved conditions for the aerobic reductive decolourization of azo dyes by Candida zeylanoides. Enzym Microb Technol 31:848–854. https://doi.org/10.1016/S0141-0229(02)00189-8
Russ R, Rau J, Stolz A (2000) The function of cytoplasmic flavin reductases in the reduction of azo dyes by bacteria. Appl Environ Microbiol 66:1429–1434. https://doi.org/10.1128/AEM.66.4.1429-1434.2000
Bilal M, Bagheri AR, Vilar DS, Aramesh N, Eguiluz KIB, Ferreira LFR, Ashraf SS, Iqbal HMN (2022) Oxidoreductases as a versatile biocatalytic tool to tackle pollutants for clean environment – a review. J Chem Technol Biotechnol 97:420–435. https://doi.org/10.1002/jctb.6743
Saravanan A, Kumar PS, Vo DVN, Jeevanantham S, Karishma S, Yaashikaa PR (2021) A review on catalytic-enzyme degradation of toxic environmental pollutants: microbial enzymes, J Hazard Mater 419:126451. https://doi.org/https://doi.org/10.1016/j.jhazmat.2021.126451
Wang X, Yao B, Su X (2018) Linking enzymatic oxidative degradation of lignin to organics detoxification. Int J Mol Sci 19. https://doi.org/10.3390/ijms19113373
da Silva Vilar D, Bilal M, Bharagava RN, Kumar A, Kumar Nadda A, Salazar-Banda GR, Eguiluz KIB, Romanholo Ferreira LF (2022) Lignin-modifying enzymes: a green and environmental responsive technology for organic compound degradation. J Chem Technol Biotechnol 97:327–342. https://doi.org/10.1002/jctb.6751
Kishor R, Saratale GD, Saratale RG, Romanholo Ferreira LF, Bilal M, Iqbal HMN, Bharagava RN (2021) Efficient degradation and detoxification of methylene blue dye by a newly isolated ligninolytic enzyme producing bacterium Bacillus albus MW407057. Colloids Surf B Biointerfaces 206:111947. https://doi.org/10.1016/j.colsurfb.2021.111947
Schlosser D, Höfer C (2002) Laccase-catalyzed oxidation of Mn2+ in the presence of natural Mn3+ chelators as a novel source of extracellular H2O2 production and its impact on manganese peroxidase. Appl Environ Microbiol 68:3514–3521. https://doi.org/10.1128/AEM.68.7.3514-3521.2002
Sosa-Martínez JD, Balagurusamy N, Montañez J, Peralta RA, Moreira RFPM, Bracht A, Peralta RM, Morales-Oyervides L (2020) Synthetic dyes biodegradation by fungal ligninolytic enzymes: process optimization, metabolites evaluation and toxicity assessment. J Hazard Mater 400:123254. https://doi.org/10.1016/j.jhazmat.2020.123254
Chagas EP, Durrant LR (2001) Decolorization of azo dyes by Phanerochaete chrysosporium and Pleurotus sajorcaju. Enzym Microb Technol 29:473–477. https://doi.org/10.1016/S0141-0229(01)00405-7
Verma P, Madamwar D (2002) Production of ligninolytic enzymes for dye decolorization by cocultivation of white-rot fungi Pleurotus ostreatus and Phanerochaete chrysosporium under solid-state fermentation. Appl Biochem Biotechnol - Part A Enzym Eng Biotechnol 102–103:109–118. https://doi.org/10.1385/ABAB:102-103:1-6:109
Wu FC, Tseng RL, Juang RS (2001) Enhanced abilities of highly swollen chitosan beads for color removal and tyrosinase immobilization. J Hazard Mater 81:167–177. https://doi.org/10.1016/S0304-3894(00)00340-X
Kalme S, Jadhav S, Jadhav M, Govindwar S (2009) Textile dye degrading laccase from Pseudomonas desmolyticum NCIM 2112. Enzym Microb Technol 44:65–71. https://doi.org/10.1016/j.enzmictec.2008.10.005
Kalme S, Ghodake G, Govindwar S (2007) Red HE7B degradation using desulfonation by Pseudomonas desmolyticum NCIM 2112. Int Biodeterior Biodegrad 60:327–333. https://doi.org/10.1016/j.ibiod.2007.05.006
Selvaraj V, Swarna Karthika T, Mansiya C, Alagar M (2021) An over review on recently developed techniques, mechanisms and intermediate involved in the advanced azo dye degradation for industrial applications. J Mol Struct 1224. https://doi.org/10.1016/j.molstruc.2020.129195
Eichlerová I, Homolka L, Nerud F (2006) Synthetic dye decolorization capacity of white rot fungus Dichomitus squalens. Bioresour Technol 97:2153–2159. https://doi.org/10.1016/j.biortech.2005.09.014
Sandhya S (2010) Biodegradation of azo dyes under anaerobic condition: role of azoreductase. Biodegradation of azo dyes, pp 39–57. https://doi.org/10.1007/698_2009_43
Menon S, Agarwal H, Shanmugam VK (2021) Catalytical degradation of industrial dyes using biosynthesized selenium nanoparticles and evaluating its antimicrobial activities. Sustain Environ Res 31. https://doi.org/10.1186/s42834-020-00072-6
Goszczynski S, Paszczynski A, Pasti-Grigsby MB, Crawford RL, Crawford DL (1994) New pathway for degradation of sulfonated azo dyes by microbial peroxidases of Phanerochaete chrysosporium and Streptomyces chromofuscus. J Bacteriol 176:1339–1347. https://doi.org/10.1128/jb.176.5.1339-1347.1994
Cui D, Li G, Zhao M, Han S (2014) Decolourization of azo dyes by a newly isolated Klebsiella sp. strain Y3, and effects of various factors on biodegradation. Biotechnol Biotechnol Equip 28:478–486. https://doi.org/10.1080/13102818.2014.926053
Mohana S, Shrivastava S, Divecha J, Madamwar D (2008) Response surface methodology for optimization of medium for decolorization of textile dye Direct Black 22 by a novel bacterial consortium. Bioresour Technol 99:562–569. https://doi.org/10.1016/j.biortech.2006.12.033
Shanmugam BK, Easwaran SN, Lakra PRD, Mahadevan S (2017) Metabolic pathway and role of individual species in the bacterial consortium for biodegradation of azo dye: a biocalorimetric investigation. Chemosphere 188:81–89. https://doi.org/10.1016/j.chemosphere.2017.08.138
Ayed L, Khelifi E, Ben Jannet H, Miladi H, Cheref A, Achour S, Bakhrouf A (2010) Response surface methodology for decolorization of azo dye Methyl Orange by bacterial consortium: produced enzymes and metabolites characterization. Chem Eng J 165:200–208. https://doi.org/10.1016/j.cej.2010.09.018
Božić AL, Koprivanac N, Šunka P, Člupek M, Babický V (2004) Organic synthetic dye degradation by modified pinhole discharge. Czechoslov J Phys 54:958–963. https://doi.org/10.1007/BF03166514
Moharikar A, Purohit HJ, Kumar R (2005) Microbial population dynamics at effluent treatment plants. J Environ Monit 7:552–558. https://doi.org/10.1039/b406576j
Peter R, Mojca J, Primož P (2019) Genetically modified organisms (GMOs). Encycl Environ Heal 49:199–207. https://doi.org/10.1016/B978-0-444-63951-6.00481-2
Tahri N, Bahafid W, Sayel H, El Ghachtouli N (2013) Biodegradation: involved microorganisms and genetically engineered microorganisms. Biodegrad - Life Sci. https://doi.org/10.5772/56194
Saxena G, Kishor R, Saratale GD, R.N. (2020) Bharagava, Genetically modified organisms (GMOs) and their potential in environmental management: constraints, prospects, and challenges, Bioremediation Ind. Waste Environ Saf:1–19. https://doi.org/10.1007/978-981-13-3426-9_1
Menn FM, Easter JP, Sayler GS (2008) Genetically engineered microorganisms and bioremediation. Biotechnol Second Complet Revis Ed 11–12:441–463. https://doi.org/10.1002/9783527620999.ch21m
Jin R, Yang H, Zhang A, Wang J, Liu G (2009) Bioaugmentation on decolorization of CI Direct Blue 71 by using genetically engineered strain Escherichia coli JM109 (pGEX-AZR). J Hazard Mater 163:1123–1128. https://doi.org/10.1016/j.jhazmat.2008.07.067
Chang JS, Lin CY (2001) Decolorization kinetics of a recombinant Escherichia coli strain harboring azo-dye-decolorizing determinants from Rhodococcus sp. Biotechnol Lett 23:631–636. https://doi.org/10.1023/A:1010306114286
Tian F, Guo G, Zhang C, Yang F, Hu Z, Liu C, Wang SW (2019) Isolation, cloning and characterization of an azoreductase and the effect of salinity on its expression in a halophilic bacterium. Int J Biol Macromol 123:1062–1069. https://doi.org/10.1016/j.ijbiomac.2018.11.175
Sandhya S, Sarayu K, Uma B, Swaminathan K (2008) Decolorizing kinetics of a recombinant Escherichia coli SS125 strain harboring azoreductase gene from Bacillus latrosporus RRK1. Bioresour Technol 99:2187–2191. https://doi.org/10.1016/j.biortech.2007.05.027
Vikrant K, Giri BS, Raza N, Roy K, Kim KH, Rai BN, Singh RS (2018) Recent advancements in bioremediation of dye: current status and challenges. Bioresour Technol 253:355–367. https://doi.org/10.1016/j.biortech.2018.01.029
Wang B, Wang Z, Chen T, Zhao X (2020) Development of novel bioreactor control systems based on smart sensors and actuators. Front bioeng biotechnol 8:1–15. https://doi.org/10.3389/fbioe.2020.00007
Su CXH, Low LW, Teng TT, Wong YS (2016) Combination and hybridization of treatments in dye wastewater treatment: a review. J Environ Chem Eng 4:3618–3631. https://doi.org/10.1016/j.jece.2016.07.026
Rahimnejad M, Adhami A, Darvari S, Zirepour A, Oh SE (2015) Microbial fuel cell as new technology for bioelectricity generation: a review. Alexandria Eng J 54:745–756. https://doi.org/10.1016/j.aej.2015.03.031
Acknowledgements
The authors acknowledge the Ministry of Higher Education Malaysia for supporting the research with Fundamental Research Grant Scheme (FRGS) the grant (FRGS/1/2022/STG04/USM/02/7) for the opportunity to carry out the research.
Funding
This work is supported fundamental research grant (FRGS/1/2022/STG04/USM/02/7) provided by the government of Malaysia.
Author information
Authors and Affiliations
Contributions
All authors contributed to the studies based on fundamental proof of concept. Anandita and Kashif Raees performed data collection, analysis, and drafting of the manuscript under the supervision of Mohammad Shahadat and Syed Wazed Ali. Mohammad Shahadat finally edited the manuscript with the help of supporting references. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethical Approval
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Anandita, Raees, K., Shahadat, M. et al. Mechanistic Interaction of Microbe in Dye Degradation and the Role of Inherently Modified Organisms: a Review. Water Conserv Sci Eng 8, 43 (2023). https://doi.org/10.1007/s41101-023-00219-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41101-023-00219-7