Abstract
Potato virus Y (PVY) causes major yield losses in potato (Solanum tuberosum L.) crops worldwide. Over the past few decades, two new PVY strains (PVYN−Wi/N:O and PVYNTN) have become prevalent and widely distributed worldwide. In Kenya, there is limited information on the prevalence and distribution of PVY strains in potato cultivars grown in the farmer’s fields. This is a major limiting factor in making decisions for setting up a basis for PVY control. This study was therefore conducted (a) to determine the present prevalence and distribution of the different PVY strains in cultivars grown in different potato-growing counties and (b) to use this knowledge on virus prevalence and distribution in proposing a strategy for PVY control. To this end, a leaf sampling survey was conducted in 354 potato farms in five major potato-growing counties in Kenya with follow-up surveys in two of the five counties for two more seasons. Collected leaf samples were tested using DAS-ELISA, and those found to be infected with PVY were subjected to reverse transcription polymerase chain reactions (RT-PCR) to determine the PVY strains. Overall PVY prevalence was generally low (8.2%). Out of all the PVY-infected plants, PVYN−Wi/N:O was the most prevalent strain (54.9%) followed by PVYO (28.0%) and PVYNTN (17.1%), while PVYN was not detected. The PVYN−Wi/N:O strain occurred across the three seasons and across the counties but only on specific cultivars. Three cultivars were identified as PVY resistant, and the most widely grown cultivar Shangi (60.2%) had only a low PVY prevalence of 0.6%. This is an indication that this cultivar may have already been useful in minimizing virus prevalence in farmer’s fields. This report is the first prevalence survey of PVY strains and resistance; in addition, the results obtained provide a basis for a major focus on PVY resistance breeding in Kenya.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato virus Y (PVY) is a problem in all regions of the world where potato (Solanum tuberosum L.) is grown. The virus is transmitted mainly through infected seed tubers and by over 50 species of aphids in a non-persistent manner (Quenouille et al. 2013). PVY belongs to the genus Potyvirus within the family Potyviridae and has a single-stranded ( +)-sense RNA genome of about 9700 nucleotides. In an infected plant, the PVY genome is translated into one large polyprotein which is then cleaved into ten functional proteins by three proteases encoded by the virus (Green et al. 2017).
The virus occurs as different strains which can be distinguished biologically, serologically and on specific differences in their genome nucleotide sequences. Historically, the description of PVY strains started in the 1930s based on the symptoms they elicited either on potato (Solanum tuberosum L.) cultivars with or without specific PVY resistance genes like in cultivars King Edward, Pentland Crown, Desiree, Pentland Ivory and Maris Bard or the symptoms on tobacco (Nicotiana tabacum) plants like in the cultivar White Burley (Smith 1931; Jones 1990; Baebler et al. 2020). This led to the distinction of three different PVY strains called PVYO, PVYN and PVYC. Later, enzyme-linked immune-sorbent assay tests (ELISA) were developed to detect and differentiate PVY isolates either as ‘O’ or ‘N’ serotypes using specific monoclonal antibodies (Rose and Hubbard 1986; Ellis et al. 1996; Crosslin et al. 2005). Due to the limitations of ELISA in differentiating recombinant PVY strains, different types of reverse transcription polymerase chain reaction (RT-PCR) assays were developed to simultaneously detect and differentiate PVY strains. From the RT-PCR tests, the most commonly reported strains are PVYO and PVYN and more recently PVYNTN and PVYN−Wi/N:O (Nie and Singh 2002; Schubert et al. 2007; Lorenzen et al. 2006). Full or partial genome nucleotide sequence analysis is often used to confirm the genetic identity of specific PVY strains and the evolutionary relationships between and within the strain groups (Green et al. 2018). Over the years, there has been an evolutionary shift resulting from genome recombination of two of the traditionally known strains (PVYO and PVYN) to new recombinant strains, predominantly PVYNTN and PVYN−Wi/N:O (Kehoe and Jones 2016; Funke et al. 2017; Tran et al. 2022) leading to changes in prevalence of particular PVY strains in different regions in the world.
Among the different ways of managing the virus, the use of host resistance is an effective long-lasting way of PVY control. Two types of resistance, extreme resistance (ER) and hypersensitive resistance (HR), to the virus have been discovered in wild relatives of potato and have been introgressed into commercial cultivars (Quenouille et al. 2013; Gao et al. 2020). The extreme resistance genes, Rychc, Rysto and Ryadg, were sourced from Solanum chacoense, S. stoloniferum and S. tuberosum ssp. andigena, respectively (Cockerham 1943; Sato et al. 2006) and are known to be durable and effective against all known strains of PVY (Valkonen et al. 2017; Biryukova et al. 2019). Cultivars with this kind of resistance do not develop any PVY infection after greenhouse inoculation and field screening experiments, unlike susceptible ones where up to 100% infection can occur (Whitworth et al. 2009; Quenouille et al. 2013; Fulladolsa et al. 2015). The hypersensitive resistance (HR) controlled by Nytbr and Nctbr genes from S. tuberosum limits virus movement within the plant by inducing systemic necrosis in virus-infected tissues (Nie et al. 2012; Jones and Stuart 2018). This is a form of resistance which leads to little or no virus translocation to the daughter tubers from the mother plant after initial infection (Bertschinger et al. 2017). Some of the sources of this type of resistance are strain-specific, i.e., such cultivars may only be resistant to older PVY strains like PVYO and PVYN but susceptible to new recombinant strains like PVYNTN and PVYN−Wi/N:O (Jones 1990; Kehoe and Jones 2016). The effectiveness of resistances in such cultivars is usually assessed by challenging (exposure to sources of infection) the cultivars with different PVY strains (Nie et al. 2012; Funke et al. 2017; Jones and Stuart 2018).
In Kenya, potato is the second most important food crop after maize (NPCK 2017). Past PVY surveys in Kenya indicated high virus prevalence of 77.6% in 2006, 70% in 2009 and 27.0% in 2011 (Gildemacher et al. 2009; Muthomi et al. 2009; Nyamwamu et al. 2014). This could be one of the reasons for the low average potato tuber yields reported (8.7 T/ha) as compared to the expected potential yield of over 40 T/ha which can be achieved if the growing conditions are improved (Lung’aho et al. 2007; Onditi et al. 2012, 2021a; FAOSTAT 2017). Studies indicate that the type and level of resistance in potato cultivars is one of the most important factors that determine relative abundance of different PVY strains in different countries (Funke et al. 2017; Dupuis et al. 2018). In Kenya, there is only limited information about the occurrence of the strain distribution of PVY in potato and the levels of field resistance in cultivars grown by farmers (Were et al. 2013). This study was therefore conducted (a) to determine the present status of prevalence and distribution of the different PVY strains in cultivars grown in different potato-growing regions and (b) to use this knowledge on PVY strains prevalence and distribution in proposing a control strategy.
Materials and methods
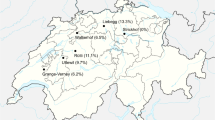
Potato leaf sampling surveys were conducted in fields of 354 ware potato farms in five of the major potato-growing counties in Kenya. These counties are among the leading potato-producing counties, and potato is the main staple food for the people in these counties. The counties are basically in the highlands of Kenya at above 1800 m.a.s.l where climatic conditions favour potato production in the two rainy seasons of the year (long rains in April to July and short rains from September to November). The first survey was conducted during the long rainy season of 2016. Two similar follow-up sampling surveys were conducted in two of the five counties (Nyandarua and Meru) using the same number of samples and on the same farms in 2016 (short rains season) and finally in 2017 (long rains season).
The geographical coordinates of the survey sites (65 farms in Nyandarua, 64 in Kiambu, 80 in Meru, 80 in Bomet and 65 in Nakuru (Fig. 1)) were generated using a randomization tool in QGIS software (QGIS 2019) according to the total number of samples for each of the five counties. Potato farms which fell on or closest to the QGIS generated GPS coordinates were identified for the survey. Leaf samples were collected from four randomly selected potato plants (without discriminating symptomatic or asymptomatic plants) within each of the 354 farms in the five counties. The four samples from each farm were tested as one composite. The samples were collected from among the first 3 leaves at the top of the potato plant. The survey was conducted at a time when the crop was at the early vegetative stage between 30 and 60 days after planting and plants had not yet started to form tubers. The samples were first subjected to double antibody sandwich enzyme-linked immune-sorbent assays (DAS-ELISA) (Clark and Adams 1977) at the Kenya Agricultural and Livestock Research Organization (KALRO), Tigoni. Samples confirmed to be infected with PVY in ELISA were subjected to a reverse transcription polymerase chain reaction (RT-PCR) at the Kenya Plant Health Inspectorate Service (KEPHIS) Laboratory, Plant Quarantine and Biosafety Station (PBQS), Muguga, to differentiate between the PVY strains. At the time of sampling, the following data were collected: the global position system (GPS), position of the farm using UTM GEO MAP (Android app), the name of the cultivar, and the PVY symptoms from the plants where the samples were collected from including scores on symptom severity where 0 = no symptom, 1 = very mild, 2 = moderately mild, 3 = moderate, 4 = severe and 5 = very severe.
A DAS-ELISA kit with antibodies and instructions was obtained from the International Potato Center (CIP), Lima, Peru. ELISA plates (Thermo Scientific, Denmark) were coated with 100 µl of coating solution consisting of 35 µl of PVY polyclonal antibodies (PVY-IgG) in 10 ml of coating buffer (0.2 g Na2CO3, 0.44 g NaHCO3, 0.03 g NaN2 in 120 ml of distilled water, pH 9.6) and incubated overnight at 4 °C. All plate washings after incubations were done in triplicate using washing buffer consisting of 8.0 g NaCl, 0.2 g KH2PO4, 0.2 g KCL, 1.13 g NaH2PO4, 0.29 NaN2 and 0.05% of Tween 20 in 1 L of distilled water, pH7.4 (PBS-T). One hundred microlitres of 1 g of leaf material macerated in 2 ml of sample extraction buffer (4.0 g PVP–40,000 and 2.0 g of egg ovalbumin in 200 ml of PBS-T) was added in duplicates to the plate and incubated overnight at 4 °C. A positive control consisting of a potato leaf sample previously tested positive for PVY in DAS-ELISA was added to three wells of each plate, and a negative control consisting of a potato leaf sample previously confirmed to be free of PVY was added to six wells of each plate. Sample loadings were conducted on multiple ELISA plates because of the large number of samples, and identical negative and positive controls were used across the plates (Khan et al. 2003). In addition, we ensured that the differences in OD values of the controls in different plates were within three times the standard deviation of their value (Khan et al. 2003). After an overnight incubation and subsequent washing, 100 µl of conjugate solution (35 µl of PVY IgG-AP in 10 ml of extraction buffer) was added and incubated for 3 h at 37 °C. After washing colour development was initiated by incubating the plates with 100 µl per well of 15 mg para-Nitrophenylphosphate (pNPP) in 20 ml substrate buffer (97 ml diethanolamine in 1000 ml distilled water, pH 9.8) at room temperature. An ELISA reader (BioTek ELX800, VT, USA) was used to take optical density (OD) values at a wavelength 405 nm after 60 min of substrate incubation. The absorbance values obtained were used to assess PVY infection in the samples as well as relative virus concentrations. Samples per individual plate were considered PVY positive when the mean OD value of the sample duplicates was equal or greater than the mean of the six negative controls plus three times the standard deviation of these negative controls (Khan et al. 2003).
All samples positive for PVY in ELISA were subjected to RT-PCR using previously described primers and protocols (Lorenzen et al. 2006; Schubert et al. 2007). Two hundred microlitres of the virus samples macerated in ELISA extraction buffer was used for total RNA extraction using TRIzol®Reagent (Life Technologies) according to the manufactures’ instructions. The dried RNA pellet was re-suspended in 30 μl of RNAse-free Milli-Q water and stored − 20 °C awaiting the subsequent cDNA synthesis.
For each individual sample, a mix consisting of 1 μl of 100 μM of each of the three reverse primers (Table 1), 2 μl of the RNA and 7 μl MQ water was incubated at 70 °C for 5 min to denature the RNA and then snap cooled in ice water. This was followed by adding 10 μl of reverse transcription mix consisting of 4 μl of 5X AMV RT buffer (Promega), 0.25 μl of 10 mM dNTP-mix, 0.5 μl of 2 mM DTT, 0.5 μl of 10u/μl AMV Reverse transcriptase (Promega) and 4.75 μl MQ water and incubating for 1 h at 42 ℃ to synthesize cDNA for the subsequent PCR process.
The PCR mixture consisted of 5 μl of 5X Taq reaction buffer (Promega), 2.5 μl of 25 mM MgSO4, 0.5 μl of 10 mM dNTP-mix, 0.5 μl of 5u/µl Taq polymerase (Promega), 8.5 μl MQ water and 1 μl of each of the six forward and reverse PCR-primers (100 μM) (Table 1) and 2 μl of cDNA. The PCR reaction was started with 2 min of denaturation at 94 °C followed by 12 cycles at 94 °C for 10 s, 30 s at 66 °C with minus 0.5 °C for each cycle and 72 °C for 60 s, followed by 20 cycles of 92 °C for 10 s, 60 °C for 30 s and 72 °C for 60 s. The PCR ended with a final extension for 72 °C for 7 min (Lorenzen et al. 2006). The amplified PCR products were separated in 1% agarose gel electrophoresis and visualized using ethidium bromide. The expected PCR products and the different fragments sizes for the different PVY strains were identified according to the distinctive fragments size as shown in Table 2 and Fig. 2.
Visualization of PCR products of each of the four potato virus Y (PVY) strains in agarose gel electrophoresis according to the expected fragment sizes (bp) for each primer combinations as summarized in Table 2. PVYN−Wi/N:O was used as a positive control
Data were analysed using IBM SPSS Statistics (2017) for Windows, Version 25.0. Armonk, NY: IBM Corp to obtain descriptive means and percentages of the occurrence of the PVY strains on cultivars in the different counties and over the three consecutive seasons.
Results
Overall prevalence of PVY strains in five potato-growing counties in Kenya
Potato leaf samples collected from ware potato fields of smallholder farmers in five major potato-growing counties in Kenya were tested for PVY infections by DAS-ELISA followed by differentiation of four PVY strains (PVYN−Wi/N:O, PVYNTN, PVYO and PVYN) in an RT-PCR. The representative agarose gel visualization of PCR products of each of the four PVY strains are presented in Fig. 2, and each of the primer combinations which gave rise to the PCR fragment sizes (bp) is presented in Table 1. Results of both the general PVY DAS-ELISA and RT-PCR strain detections are summarized in Table 3. In comparison to earlier reports (Gildemacher et al. 2009; Muthomi et al. 2009; Nyamwamu et al. 2014), the overall PVY prevalence observed in this survey was low with 8.2% of all samples being positive for PVY in DAS-ELISA. Subsequent RT-PCR analyses of the PVY positive samples showed that PVYN−Wi/N:O was the most prevalent strain (54.9%) followed by PVYO (28.0%) and PVYNTN (17.1%), while PVYN was not detected. Comparing the five counties, the highest PVY prevalence was in Meru (18.8% of positive samples) followed by 8.8% in Bomet, 7.8% in Kiambu and 3.1% in Nyandarua. PVY was not detected in the samples collected from Nakuru County.
Prevalence of PVY strains in Kenyan potato cultivars
Out of the 13 different cultivars tested, PVY was detected in only six, namely, Shangi, Kombere, Dutch Robyjn, Desiree, Nderamwana and Nyayo. Shangi, the cultivar grown by the majority of farmers (60.5%), however, had only few (3 out of 214; 1.4%) of its samples infected with PVY (0.9% PVYN−Wi/N:O and 0.5% PVYNTN). Almost a quarter (27.3%) of the samples from Kombere, the second most popularly grown cultivar (15.5%) were found with PVY infection, and the most prominent strains in the cultivar were PVYN−Wi/N:O (25.5%), PVYNTN (3.6%) and PVYO (9.1%). Dutch Robyjn was the third most popularly grown cultivar (4.0%), and half (50.0%) of the samples of this cultivar was PVY infected. The three PVY strains found in this cultivar included PVYN−Wi/N:O (42.9%), PVYNTN (14.3%) and PVYO (21.4%). The rest of the PVY-infected cultivars, Desiree, Nderamwana and Nyayo, were each grown by very few farmers, 2.0%, 0.6% and 0.8% respectively, though a larger proportion of each (14.2%, 50.0% and 66.7%, respectively) were found to be infected only with the PVYN−Wi/N:O strain. PVY was not detected in seven cultivars (Sherekea, Asante, Kenya Karibu, Tigoni, Kalucy, Unica and Thimathuti) by neither DAS-ELISA nor RT-PCR (Table 3).
Symptoms of PVY strains in potato cultivars
PVYN−Wi/N:O infections in cultivar Shangi were symptomless, but those infected with PVYNTN had chlorosis and necrotic spots on the stem (Fig. 3). All infections of PVYN−Wi/N:O, PVYNTN and PVYO were found with mosaic symptoms in cultivar Kombere (Fig. 4). Cultivars Nderamwana and Nyayo were infected only with PVYN−Wi/N:O and had mosaic symptoms. Dutch Robyjn was found with PVYN−Wi/N:O, PVYNTN and PVYO infections, and the symptoms observed included mosaic and crinkled leaves, stunted growth and leaf and stem necrosis. PVYN−Wi/N:O/N:O was the only PVY strain in cultivar Desiree with both mosaic and crinkled leaves. Mean DAS-ELISA OD values in PVY-infected samples were lowest in cultivar Shangi (0.419), the most popularly grown cultivar. The OD values were higher in Desiree (1.048), Nyayo (1.427), Nderamwana (1.443) and Dutch Robyjn (1.490), grown only by few farmers. The mean OD values were highest in Kombere (1.949), the second most popularly grown cultivar.
Possible tuber symptoms could not be assessed as by the time of sampling (about 60 days after planting) plants had not yet started to form tubers.
Occurrence of multiple infections PVY strains in potato cultivars
Out of the PVY-infected samples analysed, mixed infections of PVY strains occurred only in cultivars Kombere and Dutch Robyjn (Fig. 5). In Kombere, 6.7% of PVY-infected samples had mixed infections with PVYN−Wi/N:O, PVYNTN and PVYO, 6.7% with PVYNTN and PVYO and 20.0% with a mixture of PVYN−Wi/N:O and PVYO. A strain mixture of PVYN−Wi/N:O, PVYNTN and PVYO was found in 28.0% of the PVY positive samples in cultivar Dutch Robyjn. There were no mixed infections found in cultivars Desiree, Nderamwana, Nyayo and Shangi.
Prevalence of PVY strains in cultivars grown within each county
The prevalence of the different PVY strains was analysed in the different regions in relation to the cultivars grown (Table 4). Among all the samples analysed, the PVYN−Wi/N:O had the highest overall prevalence of 7.5% in Dutch Robyjn in Bomet, 1.6% in Shangi in Kiambu, 17.5% in Kombere in Meru and 1.5% both in Shangi and Nyayo in Nyandarua. PVYO was the second most prevalent strain, but it was found only in Kombere (6.3%) in Meru and in Dutch Robyn (3.8%) in Bomet. PVYNTN was the least prevalent PVY strain with 2.5% in Dutch Robyn in Bomet, 1.5% in Shangi in Kiambu and 2.5% in Kombere in Meru.
PVY infections were found only in particular cultivars grown across different counties. In Meru County, six cultivars were grown, but PVY was found only in Kombere, the cultivar grown by the majority (67.5%) of the farmers in this county. This cultivar had the highest overall PVY prevalence (18.8%) and also the highest prevalence of the three PVY strains (17.5% PVYN−Wi/N:O, 2.5% PVYNTN and 6.3% PVYO). Dutch Robyjn grown by 13.8% of the farmers in Bomet County was the only cultivar found infected with PVY among the four cultivars grown in the county. PVY prevalence in this cultivar was 8.8% (with 7.5% PVYN−Wi/N:O, 2.5% PVYNTN and 3.8% PVYO). In Nyandarua County, the majority of farmers (95.4%) grew cultivar Shangi, and PVY prevalence in this cultivar was very low (1.5%). PVY was also found in cultivar Nyayo (1.5%), and only PVYN−Wi/N:O was detected in these two cultivars. Kiambu County had the highest number of cultivars (11 cultivars), but Shangi was the main cultivar grown by more than a half (53.1%) of the farmers. PVY infections were found only in three cultivars, Shangi (3.1%), Nyayo (1.5%) and Nderamwana (1.6%). No PVY was detected in any of the samples collected from Nakuru County.
Prevalence of PVY strains over three consecutive growing seasons
The prevalence of PVY strains in cultivars were followed over three consecutive potato-growing seasons during the long rains 2016, short rains 2016 and long rains 2017 in Nyandarua and Meru County (Table 5). There was an overall PVY prevalence of 3.1%, 1.5% and 4.6% in Nyandarua County and 18.8%, 16.3% and 20.0% in Meru County, respectively. The same cultivars were grown in the same farms in Nyandarua County, while in Meru County, some farmers changed the cultivars they grew over the seasons. Shangi was the most predominant cultivar (95.4%) in farms in Nyandarua County, while the other three cultivars Nyayo, Unica and Tigoni were each grown only in 1.5% of the farms. Cultivar Kombere was grown in the majority of the farms (63.8%, 65.0% and 71.3% over the three seasons, respectively) in Meru County. Shangi was also grown in Meru but as the second most popular cultivar with 12.5% in the first and the second seasons and 11.3% in the third season. PVY was not detected both in DAS-ELISA and in RT-PCR in cultivars Unica, Sherekea, Tigoni, Desiree and Asante in all the samples analysed in the two counties and over the three seasons. These results are in line with the results obtained for the five different counties in the first long rains season of 2016.
Over the three seasons and across the two counties, PVYN−Wi/N:O was the most prevalent strain in all cultivars. Its prevalence was highest in Meru County with 17.5%, 16.3% and 20.0% during the first, second and third seasons, respectively. Similarly, PVYN−Wi/N:O was the most prevalent in Nyandarua during the first (3.1%) and the second seasons (1.5%), but the strain disappeared in the third season and instead PVYNTN was detected (3.1%) though the PVY prevalence in this county was lower.
Discussion
A survey was conducted in five major potato-growing counties in Kenya on randomly collected leaf samples from 354 ware potato farms. The leaf samples were tested in DAS-ELISA to identify PVY-infected samples which were subsequently tested in RT-PCR to differentiate four PVY strains (PVYN−Wi/N:O, PVYNTN, PVYO and PVYN). Follow-up surveys were conducted for two more seasons in two of the five counties to confirm the results during the first season. The overall PVY prevalence observed (8.2%) was lower than expected considering the high prevalence that had been reported for PVY in past surveys done in Kenya (Gildemacher et al. 2009; Muthomi et al. 2009; Nyamwamu et al. 2014). In a survey conducted in 2006 in Nakuru, Kirinyaga, Nyeri, Laikipia, Nakuru, Meru, Muranga and Nyandarua counties, a high PVY prevalence of 77.6% was reported from a DAS-ELISA test of tuber sprouts of 1000 randomly selected potato seed tubers informally sold in local markets (Gildemacher et al. 2009). A few years later in 2009, a prevalence of 70.0% was reported in potato leaf samples randomly collected (without bias on symptomatic and asymptomatic plants) from Meru, Nyeri, Nakuru, Narok and Nyandarua counties (Muthomi et al. 2009). A more recent survey conducted in 2011 (Nyamwamu et al. 2014) reported 27.0% PVY prevalence (on symptomatic plants), clearly lower in comparison to the two previous surveys. As observed in the present study and in previous reports, there seemed to have been a decreasing trend of PVY prevalence over the past 10 years. It is however difficult to make a direct comparison in PVY prevalence because each of the surveys was conducted differently.
Levels of PVY resistance in the cultivars grown by farmers are among the major factors determining changes in prevalence and geographical distribution of the different PVY strains (Dupuis et al. 2018). In Kenya, there is evidence that Kenyan farmers have been changing cultivars over time. For instance, cultivars Nyayo and Desiree were the major cultivars grown in the 1980s and early 1990s (Crissman et al. 1993) but are now among the least grown cultivars as reported in this study. In a survey conducted in 2008, the major cultivars grown by farmers were Tigoni (20%), Nyayo (19%), Thimathuti (9%), Dutch Robyjn (7%) and Asante (6%), while Shangi was among the least popular cultivars with only 5% (MoA/GTZ/PSDA 2009). As Shangi was now found to be the most popularly grown cultivar (60.9%) and considering the low prevalence (0.6%) of PVY in this cultivar, it is possible that the popularity of this cultivar may have contributed to the decreasing PVY prevalence in Kenya over the past 10 years. The type of resistance genes that enables it to have low prevalence and also low virus titres has not yet been investigated because it is a farmer variety whose source is not known. Due to multiple desirable traits, this cultivar was evaluated and officially registered as a variety in the year 2015 by KALRO (NPCK 2017). The fact that it has gained popularity without supply of certified seed tubers for over 10 years could be an indication of its ability to withstand degeneration resulting not only from PVY but also other locally important viruses like potato leaf roll virus (PLRV), potato virus X (PVX), potato virus A (PVA), potato virus S (PVS) and potato virus M (PVM). In a recent survey for these six potato viruses (Onditi et al. 2021a; b), cultivar Shangi, together with Sherekea, Kenya Karibu and Asante were among the cultivars with lowest virus titres of each of the six viruses. This can be an indication of tolerance to multiple infections of the viruses under field conditions and ability to withstand virus degeneration.
Setting up a PVY resistance breeding program requires knowledge of the current infection rates by the most prevalent PVY strains. This study shows that these rates may differ from one county to another and from one cropping season to another. Even though PVY is known to occur in Kenya (Gildemacher et al. 2009; Muthomi et al. 2009; Were et al. 2013; Nyamwamu et al. 2014), this study provides the very first prevalence information about the different PVY strains that occur in potato in Kenya. PVYN−Wi/N:O was the most prevalent and most widely distributed PVY strain in the cultivars grown. With knowledge of virus prevalence, it is easy to identify regions with the virus problem so that virus control can be focused on such regions. For instance, in this study, cultivars identified with low virus prevalence like Shangi can be introduced to regions like Meru where Kombere a susceptible cultivar is being predominantly grown.
The RT-PCR method employed in this study (Lorenzen et al. 2006) has the limitation that it can only distinguish between four main strains, while a broader spectrum of PVY strains is known by now. The method can in particular not distinguish between PVYN−Wi and PVYN:O. For this reason, we refer to these strains as PVYN−Wi/N:O. For future surveys, it would be advisable to use more modern methods to differentiate PVY strains such as described in Tran et al. (2022).
Screening potato cultivars for virus prevalence surveys can be a cost-effective way of virus control. Cultivars identified with low virus prevalence can be replanted over seasons resulting in lower rates of degeneration. This can be a fast way of identifying resistant cultivars and responding to the virus problem as compared to conventional breeding for virus resistance which normally takes approximately 5–10 years. The results obtained from PVY prevalence surveys are easier to implement because cultivars identified to be resistant like Shangi are already being grown by farmers. It will therefore be easier to advice farmers to select and adopt resistant cultivars from among the cultivars in their farms or from their neighbours. In addition, exotic or new improved cultivars introduced to the farmers often risk rejection if they lack one or more traits considered important by the farmers (Kaguongo et al. 2008). Considering the still very limited supply of certified seed tubers currently available through official channels in Kenya screening for PVY resistance from virus prevalence can be a viable option for addressing the PVY problem in Kenya. A follow-up study in which the main Kenyan cultivars were subjected to controlled PVY resistance screenings both in the greenhouse and the field validated the results obtained in this study (Onditi et al. 2021c). Shangi was in both cases found with very low prevalence, while other resistant cultivars like Sherekea, Kenya Karibu and Unica were found without any PVY infection. This kind of information can be useful in potato variety development and also in further validating the results obtained in this study.
When a potato plant gets exposed to high virus pressure from different sources of virus inoculum under natural growing conditions, for several seasons, without becoming infected or showing only very low infection levels of the virus, this can be regarded as a form of tolerance/resistance to that virus under such conditions (Davies et al. 1975). This study identified cultivars which were not found with any of the four PVY strains in all the five counties and over different growing seasons: cultivars Tigoni, Unica, Thimathuti, Sherekea and Asante. These cultivars however were grown only by a very limited number of farmers (0.3–4%, see Table 3) resulting in limited numbers of samples for these cultivars, which hampers clear conclusions of the true prevalence of PVY in these cultivars. However, multiple season PVY-resistance studies in the screen-house and field (Onditi et al. 2021c) showed that cultivars Sherekea and Unica were fully resistant to PVYN−Wi/N:O, while cv’s Asante and Tigoni were tolerant for PVYN−Wi infections with only low levels of virus and absence of any typical virus symptoms, supporting the observations from this study. Cultivar Shangi grown by a majority of the farmers (60.5%) and with a very low PVY prevalence of 0.6% and with very low virus titres can be used to minimize virus prevalence in the field considering that it is already well adopted among farmers.
The PVY resistance as deduced from PVY prevalence’s can routinely be used in PVY resistance screening if the results obtained are validated in controlled PVY sap inoculation experiments followed by field exposure trials (Davies et al. 1975; Solomon-Blackburn and Barker 2001). Under field conditions, PVY resistance genes protect the plants by triggering local or systemic necrosis of virus-infected tissues to eliminate initial infection or to limit cell to cell movement of the virus within the plant and to tubers, sometimes resulting in no or less infected daughter tubers as compared to the mother tubers (Bertschinger et al. 2017). In the field, a cultivar can also be unattractive to the aphid vectors making it more difficult to be infected under natural conditions (Flanders et al. 1992). When such plants are tested in ELISA, very low prevalence of the virus or very low virus titres are detected just like in the case of Shangi and other cultivars which were not detected with PVY in this study.
From this study, the main challenge of drawing conclusions about resistance of cultivars based on prevalence surveys conducted the following challenges were experienced: (a) firstly in some Counties, farmers grew mainly one cultivar (Shangi); therefore, the rest of the cultivars were underrepresented there, and (b) secondly, the overall PVY prevalence was low (8.2%). We were however confident of the data obtained with regard to effectiveness of resistance in Shangi because high number of samples (214) were analysed for this cultivar. When setting up this study, we did not expect a low PVY prevalence and instead expected high PVY prevalence because of the high prevalence observed in the past surveys (Gildemacher et al. 2009; Muthomi et al. 2009; Nyamwamu et al. 2014) and also because very few farmers have been planting certified seed tubers (5%), a predisposing factor to high virus prevalence.
We therefore recommend that prior to conducting a survey with the objective of getting knowledge about resistance in cultivars, (a) it should be first confirmed from recent surveys that there is high prevalence of the virus in question and not rely on past surveys in making decisions when setting up such an experiment, (b) it should be confirmed if the cultivars grown are almost equally represented in all farms or regions, and that (c) larger numbers of representative samples be collected to ensure a statically more sound comparison of resistance between the cultivars sampled.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Prior to conducting the study, consents of the respondents (farmers or their appropriate representatives) in the survey were sought. They were made to understand that their participation in this research activity was voluntary and that they were free to withdraw from the activity without any condition. They were also made to understand that that the data obtained (personal information) from the study will be treated with confidentiality.
References
Baebler Š, Coll A, Gruden K (2020) Plant molecular responses to potato virus Y: a continuum of outcomes from sensitivity and tolerance to resistance. Viruses 12:217–222
Bertschinger L, Bühler L, Dupuis B, Duffy B, Gessler C, Forbes GA, Keller ER, Scheidegger UC, Struik PC (2017) Incomplete infection of secondarily infected potato plants, an environment dependent underestimated mechanism in plant virology. Frontiers in Plant Science 3:8–74
Biryukova VA, Shmiglya IV, Zharova VA, Beketova MP, Rogozina EV, Mityushkin AV, Meleshin AA (2019) Molecular markers of genes for extreme resistance to potato virus Y in Solanum tuberosum L. cultivars and hybrids. Russian Agricultural Sciences 45:517–523
Clark MF, Adams AW (1977) Characteristics of the microplate method of enzyme-linked immunosorbent assay (ELISA) for detection of plant viruses. Journal of General Virology 34:475–483
Cockerham G (1943) Potato breeding for virus resistance. Annals of Applied Biology 30:105–108
Crissman CC, Crissman LM, Carli C (1993) Seed potato systems in Kenya: a case study. International Potato Centre, Lima
Crosslin JM, Hamm PB, Shiel PJ, Hane DC, Brown CR, Berger PH (2005) Serological and molecular detection of tobacco veinal necrosis isolates of potato virus Y (PVYN) from potatoes grown in the Western United States. American Journal of Potato Research 82:263–269
Davies HT, McEwen HL, Dixon NC (1975) Field testing potatoes for resistance to leaf roll and virus Y. American Journal of Potato Research 52(5):151–155
Dupuis B, Bragard C, Schumpp O (2018) Resistance of potato cultivars as a determinant factor of potato virus Y (PVY) epidemiology. Potato Research 62:123
Ellis P, Stace-Smith R, Bowler G, Mackenzie DJ (1996) Production of monoclonal antibodies for detection and identification of strains of potato virus Y. Canadian Journal of Plant Pathology 18:64–70
FAOSTAT (2017) FAOSTAT. Commodities by country, Kenya, potatoes, 2017. Available online: http://www.fao.org/faostat/en/#rankings/commodities_by_country. Accessed in August 2019
Flanders KL, Hawkes JG, Radcliffe EB, Lauer FI (1992) Insect resistance in potatoes, sources, evolutionary relationships, morphological and chemical defences, and ecogeographical associations. Euphytica 61:83–111
Fulladolsa AC, Navarro FM, Kota R, Severson K, Palta JP, Charkowski AO (2015) Application of marker assisted selection for potato virus Y resistance in the University of Wisconsin Potato Breeding Program. American Journal of Potato Research 92:444–450
Funke CN, Nikolaeva OV, Green KJ, Tran LT, Chikh-Ali M, Quintero-Ferrer CRA, Frost KF, Hamm PR, Olsen N, Pavek MJ, Gray SM, Crosslin JM, Karasev AV (2017) Strain-specific resistance to potato virus Y (PVY) in potato and its effect on the relative abundance of PVY strains in commercial potato fields. Plant Disease 101:20–28
Gao F, Kawakubo S, Ho SY, Ohshima K (2020) The evolutionary history and global spatio-temporal dynamics of potato virus Y. Virus Evolution 6:56–66
Gildemacher P, Demo P, Barker I, Kaguongo W, Woldegiorgis G, Wagoire W, Wakahiu M, Leeuwis C, Struik PC (2009) A description of seed potato systems in Kenya, Uganda and Ethiopia. American Journal of Potato Research 86:373–382
Green KJ, Brown CJ, Gray SM, Karasev AV (2017) Phylogenetic study of recombinant strains of potato virus Y. Virology 507:40–52
Green KJ, Brown CJ, Karasev AV (2018) Genetic diversity of potato virus Y (PVY): sequence analyses reveal ten novel PVY recombinant structures. Archives of Virology 163:23–32
Jones RAC (1990) Strain group specific and virus specific hypersensitive reactions to infection with potyviruses in potato cultivars. Annals of Applied Biology 117:93–105
Jones RAC, Stuart JV (2018) Strain-specific hypersensitive and extreme resistance phenotypes elicited by potato virus Y among 39 potato cultivars released in three world regions over a 117-year period. Plant Disease 102:185–196
Kaguongo WP, Gildemacher P, Demo P, Wagoire W, Kinyae P, Andrade J, Forbes G, Fuglie K, Thiele G (2008) Farmer practices and adoption of improved potato varieties in Kenya and Uganda. International Potato Centre (CIP), Lima
Kehoe MA, Jones RAC (2016) Improving potato virus Y strain nomenclature: lessons from comparing isolates obtained over a 73-year period. Plant Pathology 65(2):322–333
KEPHIS (2016) Seed potato production and certification guidelines. KEPHIS, Nairobi
Khan MS, Hoque MI, Sarker RH, Muehlbach HP (2003) Detection of important plant viruses in in vitro regenerated potato plants by double antibody sandwich method of ELISA. Plant Tissue Culture 13:21–29
Lorenzen JH, Piche LM, Gudmestad NC, Meacham T, Shiel P (2006) A multiplex PCR assay to characterize potato virus Y isolates and identify strain mixtures. Plant Disease 90:935–940
Lung’aho C, Lemaga B, Nyongesa M, Gildermacher P, Kinyae P, Demo P, Kabira J (2007) Commercial seed potato production in eastern and central Africa. Kenya Agricultural Research Institute, Nairobi
MOA/GTZ-PSDA (2009) National Potato Taskforce Report 2009. Final report. Ministry of Agriculture (MoA) and Germany Technical Cooperation (GTZ)-private Sector Development Agency (PSDA), Nairobi
Muthomi JW, Nyaga JN, Olubayo FM, Nderitu JH, Kabira JN, Kiretai SM, Aura JA, Wakahiu M (2009) Incidence of aphid transmitted viruses in farmer-based seed potato production in Kenya. Asian Journal of Plant Sciences 8(2):166–171
Nie X, Singh RP (2002) A new approach for the simultaneous differentiation of biological and geographical strains of Potato virus Y by uniplex and multiplex RT-PCR. Journal of Virology Methods 104:41–54
Nie B, Singh M, Murphy A, Sullivan A, Xie C, Nie X (2012) Response of potato cultivars to five isolates belonging to four strains of Potato virus Y. Plant Disease 96:1422–1429
NPCK (2017) Potato variety catalogue. National Potato Council of Kenya, Nairobi, Kenya. Available online: http://npck.org/Catalogues/NPCK%20CATALOGUE%202017%20booklet%20K2.pdf. Accessed in November 2019
Nyamwamu P, Mukoye B, Osogo A, Omuse C, Ajanga S, Were HK (2014) Distribution and biological characterization of potato virus Y in Kenya. Journal of Agri-Food and Applied Sciences 2:258–264
Onditi JO, Nderitu SWK, Landeo JA, Abong’ GO, Sikinyi EO, Kabira JN (2012) Release of three improved varieties for the expanded potato market in Kenya. Agriculture and Biology Journal of North America 3(5):192–197
Onditi J, Nganga N, Nyongesa M, van der Vlugt R (2021a) Farmer knowledge in potato virus epidemiology and control in Kenya. Potato Research 64:489–513
Onditi J, Nyongesa M, van der Vlugt R (2021b) Prevalence, distribution and control of six major potato viruses in Kenya. Tropical Plant Pathology 46:311–323
Onditi J, Nyongesa M, van der Vlugt R (2021c) Screening for PVYN-Wi resistance in Kenyan potato cultivars. Potato Research 64:469–488
Quenouille J, Vassilakos N, Moury B (2013) Potato virus Y: a major crop pathogen that has provided major insights into the evolution of viral pathogenicity. Molecular Plant Pathology 14:439–452
Rose DG, Hubbard AL (1986) Production of monoclonal antibodies for the detection of Potato virus Y. Annals of Applied Biology 109:317–321
Sato M, Nishikawa K, Komura K, Hosaka K (2006) Potato virus Y resistance gene, Rychc, mapped to the distal end of potato chromosome 9. Euphytica 149:367–372
Schubert J, Fomitcheva V, Sztangret-Wiśniewska J (2007) Differentiation of potato virus Y strains using improved sets of diagnostic PCR-primers. Journal of Virology Methods 140:66–74
Smith KM (1931) Composite nature of certain potato viruses of the mosaic group. Nature 127:702
Solomon-Blackburn RM, Barker H (2001) A review of host major-gene resistance to potato viruses X, Y, A and V in potato, genes, genetics and mapped locations. Heredity 86:8–16
Tran LT, Green KJ, Rodriguez-Rodriguez M, Orelanna GE, Funke CN, Nikolaeva OV, Quintero-Ferrer A, Chikh-Ali M, Woodell L, Olsen N, Karasev AV (2022) Prevalence of recombinant strains of potato virus Y in seed potato planted in Idaho and Washington States between 2011 and 2021. Plant Disease 106:810–817
Valkonen JPT, Gebhardt C, Zimnoch-Guzowska E, Watanabe KN (2017) Resistance to Potato virus Y in potato. In: Lacomme C, Glais L, Bellstedt DU, Dupuis B, Karasev AV, Jacquot E (eds) Potato virus Y: biodiversity, pathogenicity, epidemiology and management. Springer Nature, Cham, pp 207–242
Were HK, Kabira JN, Kinyua ZM, Olubayo FM, Karinga JK, Aura J, Lees AK, Cowan GH, Torrance L (2013) Occurrence and distribution of potato pests and diseases in Kenya. Potato Research 123:12–25
Whitworth JL, Novy RG, Hall DG, Crosslin JM, Brown CR (2009) Characterization of broad spectrum potato virus Y resistance in a Solanum tuberosum ssp. andigena-derived population and select breeding clones using molecular markers, grafting, and field inoculations. Am J Potato Res 86:286–296
Acknowledgements
We thank the Kenya Agricultural and Livestock Research Organization (KALRO), Kenya, and Wageningen University and Research (WUR), The Netherlands, for supporting and hosting this study. Special thanks to Mrs. Florence Munguti and the staff at the Kenya Plant Health Inspectorate Service for hosting Laboratory diagnosis of plant samples. We finally thank the Kenyan farmers for allowing us into their farms to conduct this study.
Funding
This study is financially supported by the Wageningen University and Research (WUR) and Kenya Agricultural and Livestock Research Organization as part of a collaborative PhD research project at WUR and KALRO.
Author information
Authors and Affiliations
Contributions
This work was conceptualized and discussed by all the three co-authors prior to implementation. The actual research work (experimental design, sample and data collection in the field, laboratory tests and data analysis and presentation) were done by the first author (Dr. John Onditi). Supervision and provision of equipment and facilities for conducting the experiments were done equally by the other two authors. The corresponding author (Prof. Dr.ir. René van der Vlugt) played the overall supervisory role in the project. The first draft of this manuscript was prepared by Dr. John Onditi. This was improved twice with equal contribution of all the authors before submission.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Onditi, J., Nyongesa, M. & van der Vlugt, R. Prevalence, distribution and control of potato virus Y (PVY) strains in Kenyan potato cultivars. Trop. plant pathol. 47, 659–671 (2022). https://doi.org/10.1007/s40858-022-00520-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-022-00520-1