Abstract
Purpose
Carpal tunnel syndrome is one of the common peripheral neuropathies. For magnetic resonance imaging, segmentation of the carpal tunnel and its contents, including flexor tendons and the median nerve for magnetic resonance images is an important issue. In this study, a convolutional neural network (CNN) model, which was modified by the original DeepLabv3 + model to segment three primary structures of the carpal tunnel: the carpal tunnel, flexor tendon, and median nerve.
Methods
To extract important feature maps for segmentation of the carpal tunnel, flexor tendon, and median nerve, the proposed CNN model termed modified DeepLabv3 + uses DenseNet-121 as a backbone and adds dilated convolution to the original spatial pyramid pooling module. A MaskTrack method was used to refine the segmented results generated by modified DeepLabv3 + , which have a small and blurred appearance. For evaluation of the segmentation results, the average Dice similarity coefficients (ADSC) were used as the performance index.
Results
Sixteen MR images corresponding to different subjects were obtained from the National Cheng Kung University Hospital. Our proposed modified DeepLabv3 + generated the following ADSCs: 0.928 for carpal tunnel, 0.872 for flexor tendons and 0.785 for the median nerve. The ADSC value of 0.8053 generated the MaskTrack that 0.22 ADSC measure were improved for measuring the median nerve.
Conclusions
The experimental results showed that the proposed modified DeepLabv3 + model can promote segmentations of the carpal tunnel and its contents. The results are superior to the results generated by original DeepLabv3 + . Additionally, MaskTrack can also effectively refine median nerve segmentations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The carpal tunnel is a passageway in the wrist formed by the carpal bone and the transverse carpal ligament. A diagram of the carpal tunnel (e.g. Fig. 1) is bounded by the transverse carpal ligament on the volar side and eight carpal bones on the dorsal side. The carpal tunnel contains nine flexor tendons and a median nerve that extends from the forearm into the hand. Carpal tunnel syndrome (CTS) is the most frequently encountered type of peripheral compression neuropathy, which is CTS is characterized by median nerve entrapment at the wrist, resulting in median nerve dysfunction. This phenomenon results in a thickened transverse carpal ligament, fibrotic changes of the subsynovial connective tissue, and a narrowed space of the carpal tunnel. This causes compression or entrapment of the median nerve, which further leads to variable hand pain and paralysis [1]. Medical information regarding soft-tissue interactions within the carpal tunnel can be obtained from magnetic resonance imaging (MRI). Carpal tunnel segmentation from MRI images remained an important evaluation of CTS [2]. Presently, manual segmentation is the most commonly used approach for sketching the structures of flexor tendons and the median nerve, through it is time-consuming and operator-dependent.
Structure of the carpal tunnel containing nine flexion tendons and the median nerve [1]
MRI had been widely used to diagnose CTS, which has made valuable contributions to accurate predictions of the location and types of regions of the carpal tunnel in clinical medicine [3]. However, the carpal tunnel is surrounded by several carpal bones and tightly enclose the median nerve and flexor tendons such that segmentation of the carpal tunnel and its contents is susceptible to artifacts e.g., ambiguous boundaries of flexor tendons and the median nerve on MR images. Two different categories have been proposed for the segmentation of serial cross-section carpal MRI images: region [4] and model-based methods [5,6,7]. The region-based method only considers intensity characteristics, such as intensity homogeneity, of the target tissues in the segmentation processing, but always fail to differentiate tissue with similar intensity of regions in carpal MRI images. Model-based methods can achieve more stable segmentation due to the constraints of priori knowledge, which usually require user-intervention to put the model in a good initial condition. Until now, no adequate solution exists for automatically segmenting the flexor tendons and median nerve within the carpal tunnel.
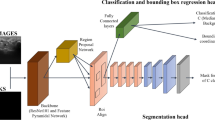
Recently, convolutional neural networks have been used to develop medical image segmentation of multimodal medical images [8, 9], which have been a widely-used method for automatic tumor segmentations of brain [10, 11], liver [12], breast [13], lung [14], rectal [15] and peripheral nerves [16]. An interesting CNN model, called the DeepLabv3 + [17], uses atrous convolution to extract the feature map at an arbitrary resolution based on the encoder-decoder structure for semantic image segmentation of a single image. Figure 2 shows the structure of DeepLabv3 + . In general, the DeepLabv3 + augments the original spatial pyramid pooling module that probes convolutional features at multiple scales by using the atrous convolution with different rates. ResNet-101[18] or Xception [19] were the backbones to extract dense feature maps by atrous convolution.
To our best knowledge, this paper represents the first attempt at fully-automatic segmentation of the flexor tendons and median nerve of the carpal tunnel from the serial cross-sectioned MRI images using a CNN. The CNN, i.e., the modified DeepLabv3 + , inputs a pair of T1 and T2 images to separate the regions of the carpal tunnel, flexor tendons and median nerve. Detail of the modified DeepLabv3 + model is shown in Sect. “Materials and Methods” Sect. “Experimental Results and Discussion” contains experimental results and associated discussions. Finally, conclusions are presented in the Sect. “Conclusion”.
2 Materials and Methods
Nine flexor tendons and one median nerve pass through the carpal tunnel in the wrist. These tissues can provide important clinical information, such as e changes in size or intensity of tissues, for measuring the severity of CTS. In this paper, a fully automatic segmentation method based on the modified DeepLabv3 + model is proposed for separating the regions of the carpal tunnel, flexor tendon, and median nerve from MR cross-section images. A flow chart of the proposed method (e.g. Fig. 3) is shown that the proposed method is divided into pre-processing, segmentation of the DeepLabv3 + , and refinement by MaskTrack and post processing. The ensemble model uses the MaskTrack method to refine the segmentation results of median nerve. In each MRI image, the segmented result of the median nerve is a complete connected component and satisfies reasonable conditions, which is selected as a reference mask, otherwise, the other results are denoted into dropped masks. Intensity of corresponding position in each dropped mask is generated by averaging the intensities of its nearest reference masks. Finally, all the results of the dropped and reference masks are integrated to establish the final segmentation results of median nerve of MRI images. Detail of ensemble model is described in Sect. 2.4.
2.1 Experimental Materials and MR mage Acquisition
The sixteen16 MR section images were obtained from the National Cheng Kung University Hospital. The instrument used was a Philips Ingenia 3.0 T MR system [20]. During imaging, subjects were asked to lie above the instrument’s platform and extend one hand forward, in the so-called superman position. Thirty-six T1 and corresponding T2 cross-section MR images of each subject in the transverse view were acquired such that the interval of adjacent slices was 2 mm in thickness. Among these slices, approximately 16 to 18 slice images contained the carpal tunnel. As shown in Fig. 4a and b, the T1 images were always sensitive to fat, such that the regions composed of fat are relatively bright; on the other hand, T2 images were sensitive to water, which serve as useful signals for identifying regions of edema. In total, 16 MR section images were captured from eight normal cases and eight patients with CTS. In the experiments, in order to efficiently train the modified DeepLabv3 + and to evaluate its performance, we indicated the start frame and the stop frame of each MR section image. The start frame is the one backward three from the distal carpal tunnel; the stop frame is the one backward three from the proximal carpal tunnel. The frames between the start and stop frames were annotated by a physician.
2.2 Data Preprocessing
Because of the different parameter settings of the acquired machine, the T1 and corresponding T2 images usually appear inconsistently in different sizes of pixel, intensity distribution, and the position of carpal tunnel. To overcome this inconsistency, several data preprocessing procedures were applied.
2.2.1 Data Normalization
Raw MR images have many of data inconsistency problems. A widely-used solution of these problems is data normalization which usually gives the training of CNN models faster convergence. Obviously, the original T1 and T2 images (e.g. Fig. 5a and b) show the lack consistency in their intensity distributions. We used Eq. (1) to adjust the intensity of each MR cross-section image.
where \({V}_{new}\) denotes the normalized factor, the \({V}_{old}\) and \({V}_{99}\) denote the original intensity and the 99 percentile of intensity distribution, respectively. The normalized intensity of each pixel is multiplied by \({V}_{new}\). The normalized results (e.g. Fig. 5c and d) look more consistent in intensity distributions. In order to normalize the pixel size, we first cropped a 100 × 100 mm region in the center of each MR image in DICOM format; then we used bilinear interpolations to resize the cropped region into 512 × 512 pixels for lateral processing.
2.2.2 Image Registration
Generally, the different weighted MR images revealed slight differences, such as in intensity and contour features. Image registration is a commonly used method to overcome this problem. A flow chart of MRI image registration is shown in Fig. 6.
A possible reason for imaging differences between T1 and T2 weighted images is that a patient’s wrists may move due to breathing as a result of the long time it often takes to obtain T2-weighted images relative to T1-weighted images. To precisely integrate the information from the T1 and T2 images, alignment of the T2 images with the corresponding T1 images was required. We registered the two different weighted MR images by using the affine transformation, which maximizes the correlation between the T1 image and the transformed T2 image based on the gradient descent method.
Intensity of the flexor tendon is always much less than the surrounding tissue in both T1 and T2 images; thus, the flexor tendon region could be roughly extracted by a single threshold. Based on experimental experience, we set the threshold to 0.125 for both T1 and T2 images. Our proposed method automatically selects a region of the flexor tendon rather than full image for establishing the correspondence.
The normalized cross correlation (NCC) is defined as the fitness function for the registration between the regions of T1 and T2 MR images. Equation (2) describes the normalized cross correlation, where \(S\) denotes the set of registered regions, \(x\) denotes a pixel in the T1 image, \(y\) denotes a pixel in the original T2 image, \(\overline{x }\) denotes the mean intensity of sampled points in the T1 image and \(\overline{y }\) denotes the mean intensity of sampled points in the T2 image.
A stochastic gradient decent with momentum [21], as shown in Eqs. (3) and (4), is then used to update the parameters of affine transformation, where \(\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}}{{a_{ij} }}\) denotes the updated direction. Based on experimental experience, t is the iterative number, the learning rate \(lr\) is set to 1, the momentum \(m\) is set to 0.9, and the weighting \(w\) is set to \(\left\{ \begin{gathered} 0.01,\;if\;j \in \left\{ {1,2} \right\} \hfill \\ 1,\;\;\;\;\;if\;j = 3 \hfill \\ \end{gathered} \right.\).
Equation (5) describes the affine transformation used. The \({a}_{ij}\) (i = 1,2; j = 1,2,3) is a parameter vector and is initialized with \(a_{ij} = \left\{ {\begin{array}{*{20}c} {1, if i = j } \\ {0, otherwise} \\ \end{array} } \right.\), \(T = \left[ {\begin{array}{*{20}c} 1 & 0 & { - 256} \\ 0 & 1 & { - 256} \\ 0 & 0 & 1 \\ \end{array} } \right]\) is used to transform the position of origin, \((x,y)\) is the position of the pixel in original T2 images and \(({x}^{\prime},{y}^{\prime})\) is the position after the transform.
2.2.3 Region of Interest Selection
In order to reduce unnecessary areas, the input images are cropped to a 224 × 224 region. In the training phase, the center of the carpal tunnel can be found by the ground truth, it is also assigned as the center of the cropped region. Then, the cropped images perform data augmentation by rotation, horizontally flipping, and image- intensity scaling. In the inference phase, since the new data have no labeled ground truth, the center of gravity of wrist (CGW) is first used estimate the center of carpal tunnel (CCT). A distribution map shown in Fig. 7 records the coordinate of the center of gravity for all of the training data generated. This result approximately shows that the center’s position of the carpal tunnel is above the center of the wrist. However, this estimated region of interest (ROI) may not be precise enough to cover the entire carpal tunnel region. Therefore, we used DeepLabv3 + to segment the carpal tunnel in this ROI; we also computed the center of the segmented carpal tunnel, which was used to crop a precise ROI for the final segmentation of DeepLabv3 + . Further, the center of gravity of the segmented median nerve from DeepLabv3 + was used to crop a ROI for MaskTrack.
2.3 Segmentation of Modified DeepLabv3 + Model
DeepLabv3 + is a powerful CNN model based on an encoder-decoder structure for semantic segmentation. DeepLabv3 + extended the structure of DeepLab v3 [22] by adding a simple decoder structure that merges the low and high-level features and up-samples the feature map by bilinear interpolation. An important technique performed by DeepLabv3 + is the atrous convolution, which increases the interval between the elements in the kernel to extend the field of view in a single convolution without additional calculations. The different dilated rate of atrous convolutions is shown in Fig. 8, 2 where the black squares denote the kernel of convolution.
The input frame of the modified DeepLabv3 + concatenates the T1 and its registered T2 images; the output channels of the output layer are background, carpal tunnel, flexor tendon, and median nerve. Figure 9 shows the structure of the modified DeepLabv3 + . At the latent space of the model, four different dilated rates of atrous convolution and an adaptive average pooling are used to perform spatial pyramid pooling, which is called “atrous spatial pyramid pooling” (ASPP). ASPP provides a fusion of feature maps in different field of view without additional computation. In this ASPP, the dilated rates of atrous convolution in ASPP are 1, 2, 3, and 4.
The original backbone of the DeepLabv3 + is the ResNet-101 [18] and Xception [19], in which several convolutions are replaced by atrous convolution with different dilated rates. In the modified DeepLabv3 + method, DenseNet-121 [6] is used as the backbone of DeepLabv3 + . According to the original architecture, atrous convolutions with different dilated rates were used in several dense blocks. Details of the DenseNet-121 architecture are shown in Table 1. The average pooling layers in transition layer 2 and 3 were removed to prevent information for small objects. Finally, following input of the stacked T1 and T2 images, the proposed model predicted the segmentation results, i.e., are carpal tunnel, flexor tendon, median nerve and background.
2.4 Ensemble Modeling
MaskTrack uses previously predicted results as training data for segmenting the next frame in the problem of video tracking. The current predicted results will be regarded as references in the next timestamp for segmentation. In this paper, the MaskTrack is used as a fine-tuned model to adjust the median nerve segmentation results generated by DeepLabv3 + . More precisely, the proposed modified DeepLabv + 3 is used as the main architecture; however, MaskTrack is used to refine the prediction of the median nerve generated by DeepLabv3 + .
Deep supervision [23,24,25] supervises the hidden layers of the model and can speed up its convergence and overcome the problem of vanishing gradients. DeepLabv3 + uses a large number of trainable parameters at ASPP in latent indoor space to train this model successfully. The deep supervision path was added after the 1 × 1 convolution behind ASPP, as shown in Fig. 9. The channel size of the high-level feature map was reduced to the output channel size and the resolution was up-sampled by bilinear interpolation to be the same as the input shape. Both the deep supervised output and final output were compared with the ground truth to obtain a loss; then the model was updated.
In order to further improve performance of the median nerve segmentation, a MaskTrack model was used to obtain more precise segmentation results by integrating the information of adjacency frames. The input of the MaskTrack model is a three-channel image that contains the target T1 image, corresponding T2 image, and reference mask of the median nerve.
The decision of the reference mask of each pair of T1 and registered T2 images is important in the use of MaskTrack. The decision is based on three stages as shown in Fig. 10. In the first stage, all segmentation results of the median nerve in the MRI images are collected from the modified DeepLabv3 + as candidate masks; then the mask filter is used to select appropriate candidates that meet the following three criteria: uniqueness, existence, and continuity. In order to satisfy uniqueness, the possible masks of a median nerve, which have two or more connected components are dropped, and then the sizes of the median nerve are re-calculated based on our used dataset. In general, the minimum size of the median nerve and average size of the median nerve are approximately 130 and 189 pixels, respectively. Without loss of generality of existence, if the median nerve size is less than \(100\) pixels, then the corresponding possible mask is dropped in order to meet the existence criterion. In order to satisfy the criterion of continuity, if the average of intersection of unions (IOU) of the current possible mask and its adjacency possible masks is less than \(T\), then the current possible mask will also be dropped. However, the selection of the IOU threshold is critical. If the threshold is too large, many possible masks will be dropped; in contrast, the remaining masks are not sufficiently accurate. MaskTrack generates larger mistakes when these candidates are used as the reference masks.
In experiments, the validation data are used to select the IOU threshold (Fig. 11). A comparison of the average Dice similarity coefficient (ADSC) (e.g. Fig. 11a) under different IOU thresholds of the resulting segmentation of the modified DeepLabv3 + is shown. The dropped rate (the ratio of dropped) under the different IOU thresholds (e.g. Fig. 11b) did not satisfy the afore-mentioned three criteria to the number of all possible masks. In Fig. 11, the ADSC is maximal as the IOU threshold is 0.3 and the corresponding dropped rate is approximately 0.2. It seems that the dropped rate violently increased when the IOU threshold was larger than 0.3. Therefore, we selected the IOU threshold to be 0.3 in order to filter out the worse candidate masks. In other words, the remaining candidates that meet the criteria of uniqueness, existence, and continuity are called “reference marks”, while, the other masks are called “dropped masks”.
The second stage is to refine the dropped frames of a median nerve by using the DeepLabv3 + with thr MaskTrack model. Bi-directional refinement was applied, as shown in Fig. 10. The resulting gray frames were retained as the reference mask. The gray frames represent the final results of the ensemble mode. The dropped frame needs further decision, which is based on the following mechanism. The reference mask and its T1 and registered T2 MR images are used as an input to generate the next prediction of dropped frames. If the next mask is a gray frame, the prediction will become the next reference mask for the following prediction. These green boxes are filled up by using the nearest predictions in a bi-directional manner. More precisely, one prediction is used to forwardly or backwardly fill up the predictions of green boxes until another gray box occurs. The final stage is to average the forward and backward predictions as the final results of the green frames in the ensemble model.
Training of the modified DeepLabv3 + and MaskTrack is independent. The Adam optimizer [24], which records the first derivative of gradient to smooth the training process, is used in both models. The batch size, momentum and weight decay are assigned as 24, 0.9, and 1 × 10–3, respectively. The models are trained for 300 epochs, at most. The weight of backbone that was pre-trained on ImageNet [25] is fixed before 15 epochs. In the first 100 iterations, the learning rate increased linearly from 1 × 10–5 to 1 × 10–3 and then remained stable at 1 × 10–3. At the 210th and the 270th epochs, the learning rate was divided by 10. Equation (6) describes the learning rate in the entire training time. The t is defined as the number of epochs:
Equation (7) describes the IOU loss function, where \(N\) denotes the batch size, \(C\) denotes the number of class, \({GT}_{c}\in \left\{\mathrm{0,1}\right\}\) denotes the ground truth at class \(c\), \({SR}_{c}\in [\mathrm{0,1}]\) denotes the segmentation results at class \(c\), and \(D{SR}_{c}\in [\mathrm{0,1}]\) denotes the deep supervised segmentation results at class \(c\).
2.5 Post-processing
The post-processing stage ensures the presence and continuity of the median nerve in the segmentation result. In all frames, each region of the segmented median nerve should be a connected component; thus, the longest continuous segmentation result is considered to be the correct position of the median nerve. Extending these correct positions, the disconnected component that is not continuous with the correct position in adjacency frames is moved in order to obtain the clear median nerve regions.
3 Experimental Results and Discussion
To measure the accuracy of the segmentation of our proposed method, all segmented results were compared with the ground truth, as labeled by an expert. The experimental data consist of the data from several patients and normal subjects. Four metrics for measuring segmentation were used to evaluate the performance of the proposed method, which are average recall (AR), average precision (AP), average Dice similarity coefficients (ADSC) and average Hausdorff distance (AHD), which are shown in Eqs. (8), (9), (10), and (11), respectively.
The Hausdorff distance is described in Eq. (12). \(N\) denotes the number of slice images of each patient, \(TP\) denotes the truth positive, \(FN\) denotes the false negative, FP denotes false positive, \(BGT\) denotes the boundary points of ground truth, and \(BSR\) denotes the boundary points of segmentation results. The AR, AP, and ADSC evaluate the similarity between the segmentation results and the ground truth, where a higher value indicates better segmentation. The AHD is used to evaluate the distance between the boundary, where a lower value indicates better results.
To obtain a fair comparison, a four-fold cross validation was used in each experiment. The four-fold cross validation divides all training MRI images into four folds in which three folds are used as training data and the remaining one is used to test the model and record the results. Repeat testing was conducted four times, which generated the test results of all the materials. In addition, we picked one-third of the training set as the validation set to select the parameters of the modified DeepLabv3 + model.
3.1 Ablation Experiment for Preprocessing
In order to confirm the necessity of preprocessing, different kinds of data were used as input to the model and the resulting performances were compared. The preprocessing steps include normalization and registration. For normalization, the model training and testing done with the T1 images DICOM files were compared with the model training and testing performed on the normalized T1 images. For registration, input stacked T1 and T2 images with or without alignment were compared. In addition, for multi-modality, simply input T1 images or the registered T2 images were also compared. Table 2 shows the four-fold cross-validation results of these experiments expressed as the mean and standard deviation of each metrics. DT1 denotes the DICOM format T1 images and, RT2 denotes the registered T2 images. All experiments were performed by modified DeepLabv3 + with output stride 16 and deep supervision.
The results in Table 2 show that the choice of T1 and its corresponding registered T2 image can effectively improve segmentation performance. More precisely, the ADSC measures of this choice were 0.930 (for carpal tunnel), 0.873 (for flexor tendon), and 0.767 (for median nerve). Apparently, the use of T1 and the registered T2 images is the best among the four possible choices, thus, the two weighted images were concatenated as inputs in the following experiment.
3.2 Classification Comparison by Using Different Backbones
Different backbones are implemented in order to determine the best choice of our proposed modified DeepLabv3 + model. The first method is the original U-Net; the second architecture is the Dense U-Net with DenseNet-121 as the backbone. The third architecture is DeepLabv3 + with ResNet-101 as the backbone. The final one is our proposed CNN model which is the modified DeepLabv3 + using modified DenseNet-121 as the backbone. Comparisons of the three target tissues are shown in Tables 3–5. Finally, we also compared the performances when the modified DeepLabv3 + is constructed with and without deep supervision. The results are shown in Table 6. In these tables, OS denotes the output stride of the backbone and DS denotes deep supervision.
Based on the results shown in Tables 3, 4, 5 the performance of the modified DeepLabv3 + is the best. It is superior to the original DeepLabv3 + + with the backbone of ResNet-101. The results show that out modification may provide some benefits, especially in the segmentation of the median nerve. However, the Dense U-Net did not generate better results, but it was only slightly worse as compared with DeepLabv3 + ; that is to say, the difference is not significant. In Table 6, additional deep supervision of the modified DeepLabv3 + can further improve performance. Based on these comparison, our proposed architecture surpassed many existing architectures in the task of tissue segmentation in the wrist MR images.
3.3 Classification of Ensemble Model
Table 6, reveals that the ADSC of the median nerve by using our proposed modified DeeoLabv3 + + is only 0.797. In order to improve the median nerve, the modified MaskTrack is further used to correct the segmentation of the median nerve from modified DeepLabv3 + . We used the ground truth of the first slice as the input reference mask and forwardly predicted the next slice. Each time, the segmentation results are passed to the next timestamp as the new reference mask. Table 7 shows the performances of the MaskTrack.
The MaskTrack with an additional reference mask was also used to show competitive performance of the additional reference mask. In order to easily distinguish the original MaskTrack method, the change is called the modified MaskTrack. In order to verify the performance of the modified MaskTrack method correction, the results from the modified DeepLabv3 + are compared with the refinement results by using the modified MaskTrack. Table 8 shows the metrics of our proposed ensemble model, in which the ADSC measurement of the median nerve segmentation exceeds 0.8053.
3.4 Qualitative Results
The segmentation results of several models are shown in Fig. 12a–c. The blue contours denote the ground truth; the red ones denote the segmentation results. The slices from distal to proximal in the labeled section with four slice intervals are listed from left to right. It can be observed that the segmentation results of U-Net and Dense U-Net exist in broken areas and missing slices. In the modified DeepLabv3 + , the broken situation is greatly improved. Some segmented results of the median nerve by using the modified DeepLabv3 + with the ensemble model are shown in Fig. 13, apparently, the predicts of median nerve conform with theground truth.
4 Conclusion
In this study, we proposed the modified DeepLabv3 + model to segment different tissue regions of carpal tunnel MR images, which include carpal tunnel, flexor tendon, and median nerve from the original DICOM files. By using the registered T2 images with the corresponding T1 images, the features in both images were integrated effectively into the proposed CNN model of the carpal tunnel, flexor tendons and median nerve. The resulting ADSCs were 0.928 for carpal tunnel, 0.872 for flexor tendons and 0.785 for median nerve. Finally, MaskTrack technology was applied to improve DeepLabv3 + with the backbone of a modified DenseNet-121. Segmentation of the median nerve achieved 0.805 for the measure of ADSC. In summary, the experimental results indicate that the modified DeepLabv3 + is effective for the different tissue segmentations of carpal tunnel MR images.
References
Reed, P. (2005). Sample topic: Carpal tunnel syndrome the medical disability advisor. US: Reed Group.
Simon, A. C., Franklyn, A. H., Andrew, C., Martin, C., & Anthony, B. (2002). Magnetic resonance neurography studies of the median nerve before and after carpal tunnel decompression. Jornal of Neurosurgery, 96, 1046–1051.
Shen, P. C., Chang, P. C., Jou, I. M., Chen, C. H., Lee, F. H., & Hsieh, J. L. (2019). Hand tendinopathy risk factors in Taiwan: A population-based cohort study. Medicine (Baltimore), 98(1), 13795.
Vincent, L., & Soille, P. (1991). Watersheds in digital spaces: An efficient algorithm based on immersion simulations. IEEE Transactions on Pattern Analysis and Machine Intelligence, 13(6), 583–598.
Michael, K., Andrew, W., & Terzopoulos, D. (1988). Snake: Active contour models. International Journal of Computer Vision, 28, 321–331.
Chen, H. C., Wang, Y. Y., Lin, C. C., Wang, C. K., Jou, I. M., Su, F. C., & Sun, Y. N. (2013). A knowledge-based approach for carpal tunnel segmentation from magnetic resonance images. Journal of Digital Imaging, 26, 510–520.
Kunze, N. M., Goetz, J. E., Thedens, D. R., Baer, T. E., Lawler, E. A., & Brown, T. D. (2009). Individual flexor tendon identification within the carpal tunnel: A semi-automated analysis method from serial cross-section magnetic resonance images. Orthopedic Research Review, 1, 31–42.
Guo, Z., Li, X., Huang, H., Guo, N., & Li, Q. (2019). Deep Learning-based Image segmentation on multimodal medical imaging. IEEE Transactions on radiation and plasma medical science, 3(2), 162–169.
Ghosh, S., Das, N., Dais, I., & Maulik, U. (2019) Understanding deep learning techniques for image segmentation. In 2019 CVRP.
Menze, B. H., et al. (2015). The multimodal brain tumor image segmentation benchmark. IEEE Transactions on Medical Imaging, 3(14), 1993–2004.
Pereira, S., Pinto, A., Alves, V., & Silva, C. A. (2016). Brain tumor segmentation using convolutional neural networks in MRR image. IEEE Transactions on Medical Imaging, 35(5), 1240–1251.
Christ, P. F., et al. (2016). Automatic liver and lesion segmentation in CT using the cascaded fully convolutional neural networks and 3D conditional random field. Proceedings of Medical Image Computing and Computer Assisted Intervention, 9901, 415–423.
Feng, X., Yang, J., Laine, A. F., & Angelini, E. D. (2017). Discriminative localization in CNNs for weakly-supervised segmentation of pulmonary modules. Proceedings of Medical Image Computing and Computer Assisted Intervention, 10435, 568–576.
Wang, S., et al. (2017). Central focused convolutional neural networks: Developing a data-driven model for lung nodule segmentation. Medical Image Analysis, 40, 172–183.
Wang, J., et al. (2018). Technical note: A deep learning-based auto-segmentation of rectal tumors in MR images. Medical Physics, 45(6), 2560–2564.
Baisiger, F., Steindel, C., Arn, M., Wagner, B., Grunder, L., EI-Koussy, M., Valenzuela, W., Reyes, M., & Scheidegger, O. (2018). Segmentation of peripheral nerves from magnetic resonance neurography: A fully automatic, deep learning–based Approach. Frontiers of Neurology, 9, 1–8.
Chen, L.C., Zhu, Y., Papandreou, G., Schroff, F., & Adam, H. (2018) Encoder-decoder with atrous convolution for semantic image segmentation. In 2018 ECCV.
He, K., Zhang, X., Ren, S., & Sun, J. (2016) Deep residual learning for image recognition. In 2016 CVPR. https://arxiv.org/abs/1512.03385.
Chollet, F. (2017) Xception: Deep learning with depth wise separable convolutions. In 2017 CVPR. https://arxiv.org/abs/1610.02357.
Philips Ingenia 3.0T. Available: https://www.philips.com.tw/healthcare/product/HC781342/ingenia-30t-mr-system.
Qian, N. (1999). On the momentum term in gradient descent learning algorithms. Neural Network, 12(1), 145–151.
Chen, L.C., Papandreou, G., Schroff, C., & Adam, H. (2017) Rethinking atrous convolution for semantic image segmentation. 2017, CVPR.
Chen, L. C., Papandreou, G., Kokkinos, I., Murphy, K., & Yuille, A. L. (2017). DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. Computer Vision and Pattern Recognition, 40, 834–848.
Chen, H., Dou, Q., Yu, L., & Heng, P. L. (2016). VoxResNet: Deep voxelwise residual networks for volumetric brain segmentation. Computer Vision and Pattern Recognition, 170, 446–455.
Dou, Q., Chen, H., Jin, Y., Yu, L., Qin, J., & Heng, P. A. (2016). 3D deeply supervised network for automatic liver segmentation from CT volumes. Medical Image Computing and Computer-Assisted Intervention, 9901, 149–157.
Acknowledgements
The authors thank the Ministry of Science and Technology, ROC (project numbered: MOST-109-2221-F-153-001, MOST-109-2634-F-006-012 and MOST-108-2622-8-006-0114) for supporting this work, which was clinically reviewed under Institutional Review Board No. A-ER-107-415.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, TH., Yang, CW., Sun, YN. et al. A Fully-Automatic Segmentation of the Carpal Tunnel from Magnetic Resonance Images Based on the Convolutional Neural Network-Based Approach. J. Med. Biol. Eng. 41, 610–625 (2021). https://doi.org/10.1007/s40846-021-00615-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40846-021-00615-1