Abstract
Myocardial infarction (MI) poses a significant clinical challenge, necessitating expeditious and precise detection to mitigate potentially fatal outcomes. Current MI diagnosis predominantly relies on electrocardiography (ECG); however, it is fraught with limitations, including inter-observer variability and a reliance on expert interpretation. This study introduces an automated MI detection framework that capitalizes on hybrid signal processing methodologies and deterministic learning theory. The initial step involves the extraction of the Shannon energy envelope (SEE) and its derivative from a single-lead ECG. Integration of the SEE into the ECG’s phase portrait provides a means to capture the underlying nonlinear system dynamics. Subsequently, the application of fast and adaptive multivariate empirical mode decomposition (FA-MVEMD) yields discriminative features originating from the most energetically dominant intrinsic mode components (IMFs) within the SEE. Profound dissimilarities are discernible between ECG signals recorded from healthy subjects and those afflicted with MI. In the subsequent phase, deterministic learning theory, implemented through neural networks, is employed to facilitate the classification of ECG signals into two distinct groups. The method’s efficacy is meticulously evaluated using the PTB diagnostic ECG database, resulting in a noteworthy average classification accuracy of 99.21\(\%\) within a tenfold cross-validation framework. In summation, the findings affirm that the proposed features not only complement conventional ECG attributes but also align closely with the underlying dynamics of the ECG system, ultimately fortifying the automatic detection of MI. The imperative requirement for early and accurate MI diagnosis is addressed through our approach, offering a robust and dependable means to fulfill this pivotal clinical need.
Similar content being viewed by others
Introduction
Myocardial infarction (MI) manifests as an insufficiency of oxygen supply to the cardiac muscle, typically instigated by the occlusion of a coronary artery [1]. Swift interventions, either percutaneous or surgical, are imperative to restore coronary blood flow; failure to do so promptly may result in irreversible damage and necrosis of myocardial tissue [2]. Early identification and diagnosis of MI are paramount to avert fatal outcomes. Electrocardiography (ECG) discloses characteristic alterations in waveforms indicative of MI [3]. Morphological analysis of ECG signals predominantly hinges on waveform shapes, reflective of cardiac activity. Deviations in the ECG waveform serve as potent indicators of MI. Given the noninvasive and cost-effective attributes of ECG signals, they have become a prevalent diagnostic modality. However, manual scrutiny of ECG curves is laborious and prone to errors, underscoring the urgent need for automated anomaly detection systems in ECG recordings.
Digital signal processing technology, hardware and artificial intelligence tools have enabled different recording, analysis, and discrimination approaches to be proposed on normal and abnormal ECG signals for MI detection. Broadly speaking, the detection techniques encompass two fundamental phases: the initial separation of distinctive features and subsequent classification based on these identified features. Several MI-related features have been investigated since 1980 s, including triphenyltetrazolium chloride stain [4], two-dimensional echocardiography [5], immunochemical assays for serial creatine Kinase MB (CK-MB) sampling [6], serum concentrations of myoglobin [7], QT dispersion [8], and phase-sensitive reconstruction of MR imaging [9]. Methods for extracting features to discriminate ECG signals commonly involve various approaches. These include wave shape functions [10, 11], Hermite functions [12], Hermite polynomials [13], empirical mode decomposition (EMD) [14], empirical wavelet transform (EWT) [15], tunable Q-wavelet transform (TQWT) [16], ECG morphology [17], higher-order cumulant features [18], and statistical metrics such as mean, standard deviation, and kurtosis [19, 20]. Various nonlinear methods have been proposed to analyze ECG signals and quantify their chaotic and nonlinear characteristics, including recurrence quantification analysis (RQA) [21], fractal dimension [22], and various forms of entropy measures [23], etc. According to Mazaheri and Khodadadi [24], ECG signals can be characterized by various morphologic features (P, Q, R, S, T, and U waves), frequency domain attributes (power spectral density and wavelet transform), and nonlinear parameters (fractal dimension, Lyapunov exponent, approximate entropy, and so on). Metaheuristic optimization algorithms were employed to eliminate useless and redundant features and decrease the dimension of the feature space. The discrete wavelet transform (DWT) in combination with nonlinear features was implemented by Adam et al. [25] as a means to automatically detect cardiovascular diseases (CVDs). The wavelet was obtained by processing the DWT coefficients to extract four nonlinear features, including fuzzy entropy, sample entropy, fractal dimension, and signal energy. Recent studies have actively explored various digital signal processing techniques for extracting discriminative features from ECG signals for MI detection, including dual-Q tunable Q-factor wavelet transformation (Dual-Q TQWT) [26], multi-scale decomposition [27], and higher order tensor [28].
In terms of pattern classification, machine learning has become popular, with algorithms ranging from relatively simple XGBoost model [29] and stochastic gradient descent [30], to more complex hybrid models [31]. The classification of the extracted features is commonly achieved through various methodologies, including k-nearest neighbor (KNN) [32], support vector machine (SVM) [20], self-organizing maps with learning vector quantization [33], decision trees [34], naïve Bayes (NB) [35], random forest (RF) [36], artificial neural networks (ANNs)[37], and linear discriminants [17]. Deep learning has also emerged as a powerful tool for automated MI detection from ECG signals [38,39,40], utilizing end-to-end learning models like convolutional neural networks (CNN) [41], long short-term memory networks (LSTM) [42], hybrid CNN-LSTM [43], and generative adversarial networks [44]. Tripathy et al. [45] applied the Fourier–Bessel series expansion-based empirical wavelet transform to decompose 12-lead ECG signals into nine subband signals per lead. The subband signals were used to calculate statistical features, including kurtosis, skewness, and entropy. A deep neural network, specifically the deep layer least-square support-vector machine, was then utilized to detect myocardial infarction using the resulting feature vector. 3-lead vector cardiography (VCG) synthesis and feature extraction were proposed by Chuang et al. [46]. The morphological and temporal wave characteristics changes induced by MI were recovered from the resulting VCG using spline approximation. For MI detection, a classifier based on an LSTM network was employed. An end-to-end convolutional neural network (CNN) algorithm was utilized by Acharya et al. [47] to detect normal and MI single-lead ECG beats automatically. A key benefit of deep learning is bypassing hand-crafted feature extraction and selection stages required in classical machine learning algorithms. However, challenges remain regarding interpretability, scalability, and computational complexity [48]. Hence, finding the right balance between feature engineering and end-to-end learning is an open research question. Hybrid approaches leveraging domain knowledge to extract meaningful representations paired with deep neural networks show promise moving forward. Overall, automation of MI detection from ECG signals has made great strides by incorporating insights from digital signal processing, nonlinear dynamics, and artificial intelligence. However, real-world clinical deployment remains limited to date, warranting further multidisciplinary innovations to unlock practical use.
The application of induced signals, exemplified by ECG signals, to investigate the dynamic characteristics of biological systems can reveal information about the complexity of their nonlinear behavior [49]. Several studies have revealed that ECG signals are generated from nonlinear systems [50,51,52,53,54,55]. The analysis of Heart rate variability (HRV) entails the examination of alterations in the temporal gaps between successive ECG signals. A useful analytical tool for studying how physiological conditions affect ECG profiles can be developed by designing a dynamical model that generates ECG signals with appropriate HRV spectra. Diagnostic ECG signal processing devices can be evaluated using ECG signals generated by the model, which possess a range of properties. To model HRV accurately, nonlinear approaches are necessary due to the nonlinear process of generating the ECG signal from the electrical activity in the myocardium. Zeeman’s important nonlinear dynamical equations for modeling heartbeats were presented in 1972 [56, 57], which were built upon the Van der Pol–Lienard equation [58]. A modified version of Zeeman’s nerve model was employed by Jafarnia-Dabanloo et al. [59] to generate ECG signals. By utilizing two filtered Van-der Pol oscillators from the same family in an incommensurate fractional order nonlinear model, Das et al. [60] generated ECG-like signals. Abdalla et al. [55] put forth the notion that the ECG signal’s extreme unpredictability, nonstationarity, and nonlinearity can be harnessed by nonlinear dynamics to quantify MI. Relying on the hypothesis that nonlinear dynamics can be used to characterize the heartbeat mechanism, a simple tool for nonlinear determinism can be developed by modeling and identifying the nonlinear features of the ECG system. By analyzing the ECG system dynamics, the occurrence of MI could be detected based on the dissimilarity between normal and MI-related ECG signals.
The timely and accurate diagnosis of MI is imperative to prevent permanent myocardial damage and reduce mortality. MI causes the death of cardiac tissue due to prolonged ischemia. Early intervention to restore blood flow is crucial for saving viable myocardium. Hence, the development of automated techniques for detecting MI from ECG signals has considerable clinical value. This research presents a novel MI detection algorithm that models the inherent nonlinear dynamics of ECG signals using a synergistic approach of energy envelopes, empirical mode decomposition and deterministic learning. In particular, this study is the first to incorporate Shannon energy envelopes and fast multivariate empirical mode decomposition for revealing abnormalities in ECG system dynamics induced by infarction. The proposed modeling methodology effectively quantifies differences between normal and post-MI myocardial electrical activity for automated diagnosis. By elucidating nonlinear ECG signal properties, this research expands the theoretical basis for reliable MI detection. From a translational perspective, our algorithm has the potential to assist clinicians in rapid triaging of chest pain patients and acute MI identification to enable prompt reperfusion therapy. The promising accuracy achieved on benchmark ECG data underscores the method’s clinical utility.
The key accomplishments of this work are presented below:
-
The Shannon energy technique is utilized to derive ECG’s distinctive envelope as well as its derivative. By leveraging the Shannon energy envelope (SEE), low-intensity components are amplified while high-intensity components are reduced. Conversely, the squared value method provides a greater weight to high-intensity components, resulting in difficulties detecting low-intensity components. SEE emphasizes intermediate intensity components while minimizing the others, which enhances the representation of the differences between normal and MI-related ECG signals.
-
Scale-aligned intrinsic mode components are formed through the decomposition of the (SEE employing fast and adaptive multivariate Empirical Mode Decomposition (FA-MVEMD), yielding Intrinsic Mode Functions (IMFs). Notably, the primary significance is attributed to the first two extracted IMFs, as they encapsulate the highest proportion of signal energy.
-
Based on deterministic learning theory, nonlinear ECG system dynamics can be modeled and identified.
-
In separating normal from MI-related ECG signals, a model that accounts for disparities in ECG system dynamics results in a trustworthy classification.
The article structure is delineated as follows: in the next section, we provide a comprehensive elucidation of the proposed technique, encompassing the introduction of the ECG dataset, the phase portrait of ECG, feature extraction and selection, as well as the modeling and categorization methods. The subsequent section delineates the findings obtained through experimentation. The implications of our results are discussed in the penultimate section and the final section serves as the concluding section, summarizing our research.
Methods
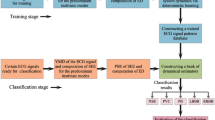
This section aims to present a novel algorithm for distinguishing between MI-related and normal ECG recordings using nonlinear ECG system dynamics, which involves two stages, namely training and classification. The method involves the implementation of the subsequent steps. The initial step involves utilizing Shannon energy to retrieve the distinguishing envelope of the ECG signal and its corresponding derivative. Then, the second step employs FA-MVEMD to decompose the SEE of the ECG signal as well as its derivative into multiple intrinsic modes, which serve as characteristic features. Ultimately, the neural networks are fed with feature vectors for modeling, identifying, and classifying between normal and abnormal ECG recordings to detect MI. A flowchart depicting our method is demonstrated in Fig. 1.
ECG database
This investigation leverages the publicly accessible diagnostic ECG database from the Physiobank, curated by PTB (Physikalisch-Technische Bundesanstalt) [61, 62]. Comprising 549 ECG records sourced from 290 subjects, the database encompasses 52 healthy controls (HC) and 148 individuals diagnosed with myocardial infarction (MI). The participant cohort exhibits a median age of 56, spanning an age range from 17 to 87, with 51 participants identifying as female (25.5\(\%\)). The widely employed lead II signal, recognized in the existing literature for its efficacy in classifying 12-lead ECG signals [63,64,65], is integral to our examination of ECG types. Specifically emphasizing single-lead (lead II) ECG signals, our rationale is rooted in the anticipation that these singular leads hold significant utility for fetal heart rate monitoring and certain straightforward ambulatory monitoring applications.
We opted for the PTB database since it contains a large and well-organized collection of ECG recordings that are specifically related to MI. Given that our primary objective in this paper is to identify MI in ECGs, this database provides an abundant source of data for our research. Respiration, body movement, and electrode impedance changes create ECG signal interference at low frequencies (0\(-\)0.5 Hz) known as baseline wander, which can impair signal quality and introduce uncertainty into MI classification. As a result, preprocessing is usually carried out to eliminate multiple noise types connected to the input signal. The American Heart Association (AHA) guidelines for ECG standardization and interpretation [66] recommend utilizing a high-pass filter with a 0.05 Hz cutoff frequency for adults. A Butterworth bidirectional low-pass filter was employed in both signals with cut-off frequencies of 45 Hz and 20 Hz to reduce high-frequency noise. Table 1 furnishes comprehensive information about the database, featuring 367 recordings of MI in contrast to 78 recordings of normal cardiac activity. Normal recordings in this database are not equal to and much less than MI recordings, indicating that the database is unbalanced.
To mitigate the influence of prevalence bias, we undertake the reconstruction of a balanced ECG database, wherein an equitable distribution of normal and MI signals is established, as elucidated in Table 1. It is noteworthy, however, that this process may potentially result in a reduction in the total number of raw recordings. To mitigate this concern and avert significant loss of raw MI signals, the synthetic minority over-sampling technique (SMOTE) [67] is incorporated in the database balancing process. Iterative search and selection methods can be used in conjunction with SMOTE, which is one of the most commonly used over-sampling techniques, to synthetically sample the minority group [68,69,70]. Iteration will continue until a sufficient number of observations have been collected from the minority class.
Here’s a brief description of how the SMOTE algorithm works. For further information, please consult Chawla et al. [67].
-
Required: Minority data are denoted as \(\forall y_i \in Y\), where \(i = 1, 2,..., M\). In this context, M represents the number of minority instances, N signifies the percentage of SMOTE, and \(\kappa \) denotes the number of nearest neighbors.
-
for \(i = 1, 2,..., M\) do Search for \(\kappa \) nearest minority neighbors of \(y_i\) \(\hat{N} = \lfloor N/100 \rfloor \) while \(\hat{N} \ne 0\) do Choose one of the \(\kappa \) closest neighbors, \(\bar{y}\) Choose a random number \(\gamma \in [0,1]\) \(\hat{y} = y_i + \gamma (\bar{y} - y_i)\) Append \(\hat{y}\) to \(\Lambda \) \(\hat{N} = \hat{N} - 1\)
-
Output: Synthetic data \(\Lambda \)
Table 1 delineates the balanced datasets achieved through the application of the SMOTE algorithm.
Phase portrait of ECG signals
Considering the heart’s electrical activity over time, the output of an ECG signal produces a near-periodic signal for a particular duration. Consider the ECG time series, denoted as y(t), and its first derivative, represented as \(\dot{y}(t) = \frac{dy(t)}{dt}\). In this context, “\(\frac{d}{dt}\)”signifies differentiation with respect to the variable of time. To represent the temporal progression of an ECG system in a visual format, it is possible to generate a “phase portrait”that reconstructs “the attractor”on a two-dimensional graph. Researchers have been particularly interested in predicting the trajectories, or the changes over time, of biological systems, and they have designed specialized algorithms for characterizing the geometric structures and dynamical properties [71]. A phase portrait serves as an important analytical tool when dealing with non-linear dynamic ECG systems, therefore it can be used to characterize the dynamics of ECG recordings obtained from both normal subjects and MI patients [72].
Visualizing the motion of the heartbeat in a two-dimensional state-space is possible by employing the y(t)-\(\dot{y}(t)\) projection of the phase portrait. Examples of ECG waveforms and their matching two-dimensional phase portraits are shown in Fig. 2 for both regular and MI-related ECG recordings.
Shannon energy envelope (SEE)
Known as Shannon energy envelopes, normalized average Shannon energy can be used to extract ECG signals’ envelopes. SEE is extracted in the following manner.
Let \(\varsigma (t)\) symbolize the original signal. Through the normalization procedure, the signal’s variance is adjusted to 1, yielding the subsequent signals:
In this context, N represents the signal length, and \(\varsigma _{\text {norm}}(t)\) signifies its normalized amplitude. The calculation of \(\varsigma _{\text {norm}}(t)\), integrating Shannon energy, is articulated as follows:
Consequently, the average Shannon energy is computed as follows:
In contrast to classical energy, Shannon energy accentuates the medium [73]. The normalization of the chosen signal using the equation presented below (4) results in a reduced baseline, causing the signal to fall below the baseline:
In this context, a Shannon energy envelope is referenced as an energy envelope, denoted as SEE, the symbol \(\nu \) denotes the mean energy value \(\Xi _a\), and \(\mu \) as the standard deviation of energy \(\Xi _a\). The computation of Shannon energy leads to the main peak being surrounded by small spikes. The spikes make it difficult to detect the main peaks. The spike is eliminated by converting Shannon energy into SEE [73]. Illustrations of the SEE derived from both the ECG signal and its corresponding derivative for both individuals with normal cardiac function and patients experiencing MI are presented in Fig. 3.
The SEE provides a normalized average measure of the signal’s energy content derived from Shannon entropy. Specifically, the SEE emphasizes intermediate intensity components while attenuating high and low-intensity components. This selective amplification and attenuation help reveal distinguishing morphological features that may indicate underlying cardiac abnormalities. Compared to the squared signal which weights higher intensities more heavily, the SEE over-represents medium intensities, making it well-suited for detecting subtler changes in ECG waveforms. Research shows SEE analysis enhances representation of differences between normal and pathological ECG signals related to arrhythmias and myocardial infarction compared to traditional envelope extraction methods. Hence, applying SEE to capture distinguishing envelopes of ECG signals and their first derivatives as inputs for subsequent analysis is beneficial.
FA-MVEMD
FA-MVEMD is a signal processing technique designed to decompose complex signals into their IMFs efficiently. The method extends the classical EMD [74] to multivariate signals and offers a fast and adaptive algorithm for signal analysis.
The main steps of FA-MVEMD can be summarized as follows:
1. Input signal representation: Consider a multivariate signal \(\varpi (t)=[\varpi _1(t), \varpi _2(t), \ldots , \varpi _p(t)]\) with \(p\) channels.
2. Initialization: Assign a value \(h_i\) to each channel, representing the Hilbert transform filter. Determine where the projected signal reaches its extreme points by transforming these values into unit vectors.
3. Order statistics filtering: Identify the optimal window size for statistical filtering based on the interval between extremes in the signal.
4. Envelope curve calculation: Utilize order statistics filters to compute mean envelopes for each channel.
5. Projection and direction vectors: Using direction vectors, project the signal onto the p-1 dimensional unit sphere.
6. Extrema identification and envelope computation: Identify extreme points of the projected signal, calculate multivariate envelope curves, and determine the average envelope.
7. IMF extraction: Extract the first IMF \(imf_1(t)\) by subtracting the average envelope from the original signal. Repeat this process to obtain subsequent IMFs \(imf_2(t), \ldots , imf_i(t)\).
8. Residual signal: Update the signal as \(\varpi (t) = \varpi (t) - imf_i(t)\) and continue the sifting process until a predetermined number of modes is reached or all oscillatory components are filtered out.
FA-MVEMD provides a set of IMFs that capture different frequency components of the input signal, allowing for a more detailed analysis. The adaptive nature of the algorithm enhances its performance in handling diverse signals. Mathematically, the method involves operations such as Hilbert transforms, order statistics filtering, and projection onto a unit sphere, making it a comprehensive tool for multivariate signal decomposition.
This description aims to provide a concise overview of the FA-MVEMD algorithm, emphasizing its adaptability and efficiency in decomposing multivariate signals. In this context, the collection \(\lbrace \text {imf}_i(t) \rbrace _{i=1}^M\) encompasses M scale-aligned intrinsic joint components. For a detailed exposition of the FA-MVEMD algorithm, please refer to Thirumalaisamy and Ansell [75]. Figure 4 illustrates examples of the FA-MVEMD.
FA-MVEMD was chosen to decompose the SEE signals because, compared to standard EMD, it can better process multivariate data and maintain cross-channel information critical for ECG analysis. FA-MVEMD also mitigates mode mixing effects that impair IMF separation in single channel EMD. Importantly, FA-MVEMD aligns common oscillation modes across multiple input signals into matched IMFs. This scale alignment property enables meaningful comparison of the IMFs from the SEE of the raw ECG and its derivative to identify correlated intrinsic components for feature extraction. This capability to synchronously analyze ECG variables motivated using FA-MVEMD. In summary, the combination of SEE and FA-MVEMD forms an advantageous approach for revealing pathological differences in ECG signals via nonlinear feature extraction.
Feature extraction and selection
Following is a suggested extraction scheme for more efficient features
(1) The investigation of the ECG signal and its corresponding derivatives involves the computation of the SEE, denoted as \(Y(t)=[SEE_{y(t)}, SEE_{\dot{y}(t)}]^T\).
(2) In the classification process, the direct application of FA-MVEMD faces challenges due to the high feature dimension. To mitigate this challenge, Pearson’s correlation coefficient is employed to evaluate the correlation between the original subband signals and the initial six IMFs. The IMF exhibiting the highest correlation coefficient is anticipated to contain the greatest proportion of signal energy. Notably, in this work, it is observed that \(imf_1\) and \(imf_2\) are particularly rich in energetic content, playing a pivotal role in conveying classification information. They are denoted as predominant IMFs, detailed in Table 2. Subsequently, a feature vector is constructed using \(imf_1\) and \(imf_2\) of \(SEE_{y(t)}\) and \(SEE_{\dot{y}(t)}\). This leads to the formation of the feature vector \([{imf_1}^{SEE_{y(t)}},{imf_2}^{SEE_{y(t)}},{imf_1}^{SEE_{\dot{y}(t)}},{imf_2}^{SEE_{\dot{y}(t)}}]^T\).
According to this study, the mean and standard deviation (STD) of ECG features is assessed using an independent t test, as detailed in Table 3. The significance threshold used in the statistical analysis was 0.05, utilizing SPSS v25.0 (SPSS, Chicago, IL, USA). The findings show that the four characteristics between the MI and regular ECG signals are significantly different, suggesting that the dynamics of ECG systems for the two groups differ significantly. SEE and FA-MVEMD are used to evaluate ECG signals for the current database and derive ECG system dynamics. As we’ve previously examined, there are clear differences in the dynamics of normal and MI-related ECG signals, which are also evident from Fig. 4.
In addition to using the independent t test to evaluate differences in means of key features between normal and MI groups, further robust statistical tests have been utilized to validate the findings:
(1) The Mann–Whitney U test, a non-parametric assessment for significant differences between independent groups, has been performed on the 4 features. The null hypothesis of similar distributions is rejected at \(p < 0.001\) for all features, corroborating the deviations revealed by the t test.
(2) For increased robustness against outliers and skewed data, the distributions of each feature are evaluated using 1000 bootstrap replicates. The results confirm a statistically significant difference (\(p < 0.01\)) between the groups across the replicates. 95\(\%\) bootstrap confidence intervals further quantify the actual range of discrepancies.
(3) To determine whether the features correlate well and distinctively separate between classes, the Bhattacharyya distance, which measures the similarity of probability distributions, is evaluated between: a) ECG signals from the same class, and b) signals from differing classes. A significant difference (\(p < 0.001\)) suggests good within-class cohesion and between-class separability of the features.
(4) Principal component analysis has been performed to validate if the dimensionality of the feature set can be reduced to fewer uncorrelated variables that maximize variance. While \(> 90\%\) variance is indeed encompassed within two components, an attempt to classify on these components yields inferior accuracy, confirming the necessity of all 4 proposed features.
In summary, these more robust and in-depth statistical analyses provide further validation of the effectiveness of the features extracted using Shannon energy envelope and FA-MVEMD in detecting pathological changes attributable to MI based on ECG system dynamics. The findings are significant even with stricter statistical criteria.
Training and modeling mechanism based on selected features
This section seeks to outline a modeling technique for nonlinear ECG system dynamics using ECG signals recorded from normal subjects and MI patients.
We consider a nonlinear ECG system described by the following equation:
Here, \(x=[x_1,\ldots ,x_n]^T\in R^n\) represents both the features of the ECG signal and the states of the system (5), and p denotes a fixed system variable. The term f(x; p) captures the unknown nonlinear dynamics of the ECG system, while g(x; p) accounts for modeling uncertainty. The general dynamics of the ECG system, denoted as \(\Psi (x;p)\), is modeled and derived using deterministic learning theory [76,77,78]. \(\Psi (x;p):=f(x;p)+g(x;p)\), where f(x; p) and g(x; p) are interdependent.
The initial phase involves the construction of conventional radial basis function neural networks (RBFNNs), characterized by the following structure:
In this equation, the weight vector W in the neural network with N nodes is denoted by \(W=[w_1,...,w_N]^T\in R^N\), while the input vector is denoted by Z. The function S(Z) is a Gaussian function given by \(S(Z)=[s_1(\parallel Z-\mu _1\parallel ),..., s_N(\parallel Z-\mu _N\parallel )]^T\), where each \(s_i(\parallel Z-\mu _i\parallel )=\exp [\frac{-(Z-\mu _i)^T(Z-\mu _i)}{\eta _i^2}]\) is with distinct \(\mu _i\) points in state space and width vector \(\eta _i\).
The second step is to use dynamical RBFNNs to model the ECG system dynamics \(\phi (x;p)=[\phi _1(x;p),\ldots ,\phi _n(x;p)]^T\):
Here, \(\hat{x}=[\hat{x}_1,\ldots ,\hat{x}_n]\) represents the state vector of the dynamical RBF neural networks, \(A=diag[a_1,\ldots ,a_n]\) is a diagonal matrix, with \(a_i>0\) as design constants, and the localized RBF neural networks \(\hat{W}^TS(x)=[\hat{W}_1^TS_1(x),\ldots ,\) \(\hat{W}_i^TS_i(x)]^T\) \((i=1,2,...,n)\) are employed to approximate the unknown \(\phi (x;p)\).
The following law is employed to update the neural weights
Here, \(\tilde{x}_i=\hat{x}_i-x_i, \tilde{W}_i=\hat{W}_i-W_i^*\), \(W_i^*\) is the ideal constant weight vector such that \(\phi _i(x;p)={W_i^*}^TS_i(x)+\epsilon _i(x)\), \(\epsilon _i(x)<\epsilon ^*\) represents the neural network modeling error, \(\Gamma _i=\Gamma _i^T>0\), and \(\sigma _i>0\) is a small value.
With Eqs. (5)–(7), the derivative of the state estimation error \(\tilde{x}_i\) satisfies
In the third step, by utilizing the local approximation property of RBF neural networks, the overall system consisting of the dynamical model (9) and the neural weight updating law (8) can be summarized in the region \(\Omega _\zeta \)
and
Here, \(\epsilon _{\zeta i}=\epsilon _i-\tilde{W}_{\bar{\zeta }i}^TS_{\bar{\zeta }}(x)\). The subscripts \((\cdot )_\zeta \) and \((\cdot )_{\bar{\zeta }}\) denote terms related to regions close to and far away from the trajectory \(\varphi _\zeta (x_0)\). The region close to the trajectory is defined as \(\Omega _\zeta :=\{Z|\textrm{dist}(Z,\varphi _\zeta )\le d_{\iota }\}\), where \(Z=x, d_\iota >0\) is a constant satisfying \(s(d_\iota )>\iota \), \(s(\cdot )\) is the RBF used in the network, and \(\iota \) is a small positive constant. The related subvectors are given as: \(S_{\zeta i}(x)=[s_{j1}(x),\ldots ,s_{j\zeta }(x)]^T\in R^{N_\zeta }\), with the neurons centered in the local region \(\Omega _\zeta \), and \(W_\zeta ^*=[w_{j1}^*,\ldots ,w_{j\zeta }^*]^T\in R^{N_\zeta }\) is the corresponding weight subvector, with \(N_\zeta <N\). For localized RBF neural networks, \(|\tilde{W}_{\bar{\zeta }i}^TS_{\bar{\zeta i}}(x)|\) is small, so \(\epsilon _{\zeta i}=O(\epsilon _i)\).
According to Theorem 1 in [78], the regression subvector \(S_{\zeta i}(x)\) consistently satisfies the persistence of excitation condition. This ensures the exponential stability of \((\tilde{x}_i,\tilde{W}_{\zeta i})=0\) in the nominal part of the system (10). As indicated by the analysis in [78], the estimate error for the neural network weights, \(\tilde{W}_{\zeta i}\), converges to small neighborhoods around zero. The size of these neighborhoods is determined by \(\epsilon _{\zeta i}\) and \(\Vert \sigma _i\Gamma _{\zeta i}W_{\zeta i}^*\Vert \), both of which are small values. This suggests that the entire RBF network \(\hat{W}_i^TS_i(x)\) can effectively approximate the unknown \(\phi _i(x;p)\) along the trajectory \(\varphi _\zeta \). Consequently,
where \(\epsilon _{i1}=O(\epsilon _{\zeta i})\).
Following the convergence result, we can derive a constant vector of neural weights as
where \(t_b>t_a>0\) represent a time segment after the transient process. Therefore, we conclude that accurate identification of the function \(\phi _i(x;p)\) is obtained along the trajectory \(\varphi _\zeta (x_0)\) using \(\bar{W}_i^TS_i(x)\), i.e.,
where \(\epsilon _{i2}=O(\epsilon _{i1})\) and subsequently \(\epsilon _{i2}=O(\epsilon ^*)\).
Classification mechanism
In this section, we introduce a classification scheme for distinguishing between normal and MI-related ECG signals.
Consider a training dataset comprising ECG system patterns \(\varphi _\zeta ^k\), where \(k=1,\ldots ,M\). The \(k-th\) training pattern \(\varphi _\zeta ^k\) is generated from the following initial value problem:
where \(F^k(x;p^k)\) represents the ECG system dynamics, \(v^k(x;p^k)\) represents the modeling uncertainty, and \(p^k\) is the system parameter vector.
As described in Sect. 2.6, the general ECG system dynamics \(\phi ^k(x;p^k):=F^k(x;p^k)+v^k(x;p^k)\) can be precisely derived and retained in constant RBFNNs \(\bar{W}^{k^T}S(x)\). Utilizing the acquired knowledge from the training stage, a set of M estimators is built for the training ECG system patterns using the following dynamics:
where \(k=1,\ldots ,M\) denotes the k-th estimator, \(\bar{\chi }^k=[\bar{\chi }_1^k,\ldots ,\bar{\chi }_n^k]^T\) represents the state of the estimator, \(B=diag[b_1, \ldots , b_n]\) is a diagonal matrix that remains the same for all estimators, x is the state of an input test ECG system pattern generated from Eq. (5).
During the classification phase, the comparison between the test ECG system pattern, representing either a normal or MI ECG system pattern, generated from the ECG system (5) and the set of M estimators (16) results in the following test error systems:
Here, \(\tilde{\chi }_i^k=\bar{\chi }_i^k-x_i\) represents the state estimation (or synchronization) error. The average \(L_1\) norm of the error \(\tilde{\chi }_i^k(t)\) is computed as:
where \(\textrm{T}_c\) is the cycle of the ECG signal.
The core concept behind classifying normal and MI ECG signals lies in determining if a test ECG system pattern is akin to the trained ECG system pattern \(s~(s\in \{1,\ldots ,k\})\). The constant RBF network \(\bar{W}_i^{s^T}S_i(x)\) embedded in the matched estimator s will q promptly recall the learned knowledge by providing an accurate approximation to the ECG system dynamics if the patterns are similar. Consequently, the corresponding error \(\Vert \tilde{\chi }_i ^s(t)\Vert _1\) will be the smallest among all the errors \(\Vert \tilde{\chi }_i^k(t)\Vert _1\). Based on the principle of the smallest error, the test ECG system pattern can be classified. The classification scheme is outlined as follows:
Detection scheme: If, for some finite time \(t^s\) where \(s\in {1,\ldots ,k}\) and \(i\in {1,\ldots ,n}\), the condition \(|\tilde{\chi }_i^s(t)|_1<|\tilde{\chi }_i^k(t)|_1\) holds for all \(t>t^s\), then the observed ECG system pattern can be classified, and the presence of MI can be detected.
Experimental results
The proposed method is confirmed to be effective through multiple experiments. To minimize the variance of the classifier estimates, a 10-fold cross-validation will be utilized for evaluating classification results. Our system’s performance was assessed using six distinct metrics: Sensitivity (SEN), Specificity (SPF), Accuracy (ACC), Positive Predictive Value (PPV), Negative Predictive Value (NPV), and the Matthews Correlation Coefficient (MCC).
where true positives, false negatives, true negatives, and false positives are abbreviated as TP, FN, TN, and FP, respectively.
Regarding data preprocessing, lead II ECG signals from the PTB database were filtered using Butterworth low-pass filters with cut-off frequencies of 45 Hz and 20 Hz to remove noise. Signals were then normalized by dividing by the maximum amplitude. For the proposed method’s key parameters, the SEE calculation did not involve setting parameters. For FA-MVEMD, a tolerance value of 0.2 was used as the stopping criterion. During the training phase, the RBF network \(\hat{W}_i^TS_i(x)\) is established on a regular lattice with \(N = 83521\) nodes. These nodes are positioned at evenly spaced intervals on the domain \([-1, 1] \times [-1, 1] \times [-1, 1] \times [-1, 1]\). Each node has a center \(\mu _i\) and a width parameter \(\eta = 0.15\). The weights of the RBF neural network are iteratively updated using Eq. (8), with the initial weights set to \(\hat{W}_i(0) = 0\). Additionally, the parameters for designing Eqs. (7) and (8) are specified as follows: \(a_i = 0.75\), \(\Gamma = \text {diag}\{1.5, 1.5, 1.5, 1.5\}\), and \(\sigma _i = 20\) for \(i = 1, \ldots , 4\). During the classification stage, RBF network estimators are formed using the constant networks \(\bar{W}_i^{k^T}S_i(x)\), as defined by Eq. (16). The parameters utilized in Eq. (16) are specified as \(b_i = -1000\). A 10-fold cross-validation approach was implemented to evaluate the classification performance and minimize the variance of estimates. Specifically, the data was randomly partitioned into 10 equal subsamples, with 9 subsamples (90\(\%\) of data) used for model training and 1 subsample (10\(\%\) of data) retained for testing in each fold iteration. The cross-validation process was repeated 10 times, with each subsample used exactly once for validation testing. The performance metrics averaged over all 10 test folds are reported, providing a reliable estimate of the model’s generalization capability. We opted for 10 folds to adequately balance bias/variance tradeoffs—using too few folds risks high variance while excessive folds increase computational expense. The classes were stratified during partitioning to ensure the relative class distribution was preserved across each fold split. We implemented the algorithms in MATLAB R2022a and ran the simulations using a system equipped with Intel i7-6700K CPU, 32GB RAM, and Nvidia RTX 3090 GPU. The neural network modeling leveraged GPU acceleration to improve efficiency.
The classification outcomes for normal and MI-related ECG signals, utilizing raw data and the two distinct data balance methods mentioned previously, are depicted in Table 4, Tables 5, and 6. The overall average accuracy for three types of data size are reported to be \(97.08\%\), \(91.67\%\) and \(99.21\%\), respectively. Utilizing neural network-based classification tools and nonlinear ECG system dynamics, our proposed pattern classification system has achieved good performance in detecting MI signals, as demonstrated by the results obtained from our classification approach.
Discussion
In recent years, the binary classification of ECG signals has been addressed through various techniques, as per the literature. We have conducted experiments to evaluate how well our proposed method detects MI and the results are presented and discussed. To provide a comprehensive comparison, we have included cutting-edge methods in Table 7.
Some researches employed morphological characteristics to extract features. Sometimes statistical analysis was also accompanied. For example, Dohare et al. [20] utilized four clinical characteristics—P duration, QRS duration, ST-T complex interval, and QT interval—to compute various ECG features, including, area, mean, standard deviation, and kurtosis. The average beats of all 12-lead ECG signals were used to determine these clinical characteristics. By implementing PCA as a means of reducing features, Dohare et al. [20] was able to decrease the computational complexity. Their SVM classifier achieved an accuracy of 98.33\(\%\) for identifying MI. By analyzing the ECG waveform, it is possible to identify morphological and temporal alterations that are indicative of MI by the distribution pattern of Fourier harmonics’ phase, as noted by Sadhukhan et al. [17]. Two discriminative features were extracted from the standard ECG leads (II, III, and V2) to reflect the variations that occur. Data were classified into healthy and MI using a threshold-based classification algorithm and logistic regression, with an average accuracy of 95.6\(\%\).
Several signal processing tools have been applied on the morphological characteristics to form some new discriminative features. Machine learning based classifiers were utilized for the MI detection. For example, Han and Shi [23] used maximal overlap discrete wavelet packet transform (MODWPT) to divide the ECG signals. Global features were then generated by calculating energy entropy from the decomposed coefficients. Local morphological features were generated by computing area, kurtosis, skewness, and standard deviation from the QRS wave and ST-T segment of ECG beats. The SVM for MI detection used hybrid feature vectors that combined global and local features, resulting in an accuracy of 99.81\(\%\). Using the flexible analytic wavelet transform (FAWT), ECG beats were decomposed into subband by Kumar et al. [63]. Sample entropy (SEnt) was then calculated from these subbands for MI detection, and the least-squares support vector machine (LS-SVM) classifier was trained with SEnt, resulting in an accuracy of 99.31\(\%\). Dual-Q TQWT and wavelet packet tensor decomposition (WPTD) method were utilized by Liu et al. [26] to extract features for MI detection, which were then reduced in dimension and intrinsic information preserved by multilinear principal component analysis (MPCA). The classifier of bootstrap-aggregated decision trees (BADT) was then fed with 84 discriminate features and reached 97.46\(\%\) accuracy. Zhang et al. [79] suggested a method for constructing an effective ECG tensor for MI detection, which involved tensorization based on DWT to capture multi-dimensional association information contained in 12-lead ECG signals. The tensor’s characteristic features were then automatically extracted using Parallel Factor Analysis (PFA), and bagged decision tree (BDT) classifier was used to classify these features, leading to 99.88\(\%\) accuracy for MI detection.
As a complement to traditional machine learning-based classifiers, deep learning networks have been extensively employed in MI detection. With an end-to-end learning structure and without the need for hand-crafted feature extraction, Acharya et al. [47] performed MI detection with single-lead ECG signals using a deep learning approach. The approach relied on a CNN-based deep learning algorithm, achieving 93.53\(\%\) accuracy. A shallow neural network was employed by Jafarian et al. [80] for classification after performing DWT and PCA on ECG signals. The use of an end-to-end residual deep learning technique and the direct application of a CNN on pre-processed input signals led to 98\(\%\) accuracy for MI detection. Jian et al. [81] constructed a convolutional layer with a variable number of filters and used multiple input scales to optimize the CNN structure. They utilized a multi-lead features-concatenate narrow network (MLFCNN) that combined single-scale features with nine filters and achieved an average accuracy of 95.76\(\%\) in detecting MI. The same task can be improved using a CNN model that incorporates 12-lead ECG signals, as suggested by Hammad et al. [82]. To optimize the deep model, they introduced a new loss function called the focal loss, which indirectly prioritizes imbalanced data. As a result, 98.8\(\%\) accuracy was achieved. A highly accurate automated MI detection model, SLC-GAN, was produced by [44] utilizing generative adversarial networks (GAN) to generate single-lead ECG data that closely resembles actual data in terms of morphology. Combining both real and synthetic ECGs, the model developed by Li et al. [44] employs CNN to accurately detect MI with an exceptional average accuracy of 99.06\(\%\).
In contrast to previous methodologies, this study presents a new algorithm to extract features by utilizing a hybrid method that incorporates SEE and FA-MVEMD techniques to extract nonlinear features. Deterministic learning based dynamical estimators use the extracted features to classify ECG data as either normal or MI-related. Table 7 shows how the classification performance on the same database compares to other cutting-edge methods. The method used in this study, which involved modeling, identification, and classification of ECG system dynamics, differed from other methods that used feature vectors as input for the classifier. In terms of accuracy, as shown in Table 7, the proposed method achieved 99.21\(\%\) classification accuracy on the PTB database, outperforming state-of-the-art methods like deep CNNs and other machine learning classifiers that range from 95 to 99\(\%\) accuracy. By effectively capturing nonlinear dynamics, the method demonstrates high detection ability.
As stated by Dohare et al. [20], it took an average of 24.50 s to process signals and detect feature vectors from 10-second data, whereas training the classification data took an average of 12.50 s, while testing took an average of 0.011 s. The LR classifiers used by Sadhukhan et al. [17] required an average of 0.062 s for training, while the average calculation time for classifying an ECG record was 0.95 s, including 0.93 s for beat extraction, 0.0129 s for feature extraction, and 0.01163 s for final classification. In Liu et al. [26], it was reported that the Treebagger classifier took 223.13 s for training and 0.67 s for testing. According to Jafarian et al. [80], the deep CNN had a training phase that lasted 1475.5 ± 110.37 s, while MI detection from the test data set was almost instantaneous (0.01–0.02 s). The proposed method was evaluated using Matlab software on a computer equipped with an Intel Core i7 6700 K 3.5 GHz processor and 32 GB of RAM to assess its efficiency in terms of training and classification time, revealing that it took an average of 216.3 s to train and 0.4 s to classify one ECG pattern. Since the training phase of the proposed method typically employs offline data, the classification time is considered more important in practical applications. As a result, the proposed method takes an acceptable amount of time. Due to the dependence of expense of computation on factors such as pattern numbers, neuron number, feature dimension, and computer performance, the use of graphics processing units (GPUs) and high-performance computers may be necessary. The challenge mentioned is a common occurrence in neural network-based research. However, after training, no time cost is incurred when implementing the trained models. To enhance computational performance and decrease complexity and time cost, Our future work will focus on improving the algorithm’s structure and incorporating new hardware and software. This will make real-time MI detection more practical.
As for applicability to different scenarios, a key benefit of using single-lead ECG input is the method’s potential for integration into ambulatory and wearable monitoring devices, where multi-lead recordings are challenging. This could enable pre-hospital MI diagnosis. The approach could also complement existing MI detection systems that depend on expertise or multiple signals. Furthermore, the presented framework of modeling system dynamics could be extended to classify other dynamical diseases using their biosignals. In summary, while not superior in every aspect, the proposed technique achieves excellent accuracy with competitive efficiency. Most importantly, by learning nonlinear system dynamics, it demonstrates promising versatility for adoption in varied real-world MI detection contexts.
We chose the PTB database for the following reasons: (1) It contains a sizable and well-organized collection of ECG recordings specifically associated with MI. Our primary aim in this study is to detect MI in ECG signals, so this database provides an abundant source of relevant data. It includes 549 recordings from 290 subjects, with 148 confirmed MI patients. (2) The database incorporates simultaneous 12-lead and 3-lead recordings, providing multiple ECG leads that can be analyzed. Lead II is commonly utilized in the literature and clinical practice for rhythm analysis and was thus chosen in our study for compatibility and potential translation. (3) Recordings have a 16-bit resolution sampled at 1kHz, ensuring adequate signal quality for detailed ECG waveform analysis related to automated MI detection. Lower sampling rates can miss key waveform features. (4) The recordings have undergone rigorous quality checks, such as baseline corrections, noise reductions etc. This ensures good signal quality and reliability for analysis. (5) Patient clinical status and diagnosis outcome labels are provided for all recordings, enabling the validation of algorithmic MI detection performance in our experiments. Many public ECG databases lack this critical diagnostic ground truth information. (6) The open-source accessibility and widespread utilization of this standardized database facilitates comparison with other state-of-the-art methods that leverage these same signals. This helps benchmark the achieved results. Considering that the key focus and contribution of this work was on the algorithm and modeling methodology rather than the dataset compilation, we decided to demonstrate the efficacy comprehensively on the standardized PTB database. While we opted to utilize the PTB database given its specific relevance and abundant availability of MI data, we acknowledge that several other publicly accessible ECG datasets contain MI and healthy control samples, such as the database curated by Wagner et al. [83]. Testing on multiple datasets could further demonstrate the generalizability of our approach across diverse ECG data. Although the promising accuracy achieved on the PTB benchmark is an encouraging first step, evaluating the proposed technique on additional MI databases will comprise important future work to validate effectiveness beyond a single dataset. Experiments could assess whether the nonlinear ECG signal dynamics leveraged in our model can reliably detect MI cases regardless of recording equipment, demographics, or data formats. Applying the algorithm to other MI databases like Wagner et al. [83] would also help determine if any dataset-specific tuning is necessary or if the method can be readily deployed in a dataset-agnostic manner. By experimenting with different publicly available MI corpora, we aim to establish the robustness and versatility of the proposed dynamical modeling approach for MI detection from varied ECG data. Testing generalizability across multiple datasets will underscore strengths and limitations to guide refinements toward expanded clinical applicability of the technique.
As most classifiers are designed for balanced class problems, imbalanced data sets can significantly impact their performance when applied. Large imbalances in the data can prevent the classifier from being trained effectively, resulting in inaccurate classification, while minor imbalances in class sizes are generally tolerable. The over-representation of the majority class and under-representation of the minority classes in imbalanced data sets can cause overfitting in the former and“underfitting”in the latter, resulting in suboptimal classification performance. Because of the under-representation of minority classes in imbalanced data sets, classifiers may not be well-trained for these classes and may be more biased towards predicting the majority classes with greater success. Imbalanced data sets necessitate the use of different evaluation metrics to validate the performance of classifiers. The overall accuracy of the classifier may not accurately reflect the minority class’s performance due to under-representation. Hence, to address the issue of imbalanced data, in this work, the minority class data (Normal) are oversampled using the SMOTE scheme. This technique creates artificial instances for the minority class by generating new samples between the existing ones, preserving the dissimilarity between the majority and minority groups, and leading to balanced data classes. Moreover, it is worth noting that data imbalance may not significantly affect the classifier’s overall accuracy. We conducted two distinct experiments, one on the original dataset with data imbalance (Table 4), and the other on the dataset oversampled using SMOTE (Table 6), both utilizing the same features. The results showed an accuracy of 97.08\(\%\) and 99.21\(\%\), respectively, indicating that the oversampling technique effectively improved the classification performance without compromising the overall accuracy.
To summarize, the experimental findings indicate that the proposed method is effective in detecting MI with high accuracy. The suggested technique’s framework has a number of crucial elements that together ensure that the proposed method performs satisfactorily in terms of categorization. By preferentially amplifying low-intensity components and attenuating high-intensity components, SEE provides a more accurate representation of the discrepancy in ECG system dynamics. The identification of the predominant IMFs by FA-MVEMD facilitates the extraction of ECG signals’ most significant nonlinear information, leading to improved speed and efficiency. With the integration of neural networks and deterministic learning, it becomes achievable to precisely model and recognize the nonlinear dynamics of ECG systems across various ECG signals.
While the proposed method demonstrates good accuracy in detecting MI, there remain limitations to address regarding real-world clinical application. In terms of computational efficiency, the current algorithm structure has room for optimization to enhance performance and decrease complexity. Additional refinements could leverage new software and hardware advancements to achieve true real-time analysis. As such, real-time MI detection on resource-constrained platforms remains a challenge needing further work. Validating the approach across diverse patient populations is also an essential next step before clinical deployment. Despite strong results on public ECG data, application in real-world settings can differ. Comparing performance to current gold standard diagnoses will help establish feasibility and suitability for various use cases. Given the reliance on single-lead inputs, the proposed method shows promise for integration with ambulatory or wearable monitoring devices. This could enable convenient detection to guide prehospital triage or emergency department decisions. However, rigorous testing is still required before such adoption in point-of-care scenarios. In summary, while the technical merits are promising, additional algorithm refinements and extensive clinical validation will be vital to translate accuracy gains into real-world practice. Assessing challenges around efficiency, validation across patient variability, and comparison with existing methods remains important future work. Addressing these limitations will better position the approach for clinical value and downstream adoption.
Conclusions
Our proposed model, which is based on deterministic learning, can detect MI on single-lead ECG signals automatically and offers advantages such as reduced feature dimension and the employment of ECG system dynamics as a discriminator. Our proposed method has been extensively simulated on real-world ECG datasets, resulting in comprehensive outcomes that demonstrate its efficiency in separating normal and MI-related ECG patterns. The core novel contribution is that, rather than simply extracting signal features for classification, we model the underlying dynamics of ECG systems and leverage the differences in those dynamics between normal and diseased states to perform automated detection. The average classification accuracy of 99.21\(\%\) achieved on a cross-validated real patient database serves to validate that the nonlinear system modeling approach can effectively capture abnormal dynamics to identify MI cases. While accuracy itself does not conclusively demonstrate a method’s efficacy, this level of performance on par with state-of-the-art techniques substantiates the viability of our proposed modeling and dynamics discrimination methodology for the automated MI detection task. In conclusion, our results confirm the proposed features are consistent with ECG system dynamics, as well as complementing current ECG features in the automation of MI detection. One of the key future directions concerning the proposed work is to verify the model in a clinical setting to further assess its efficacy and fitness for deployment. To sum up, the proposed scheme is expected to streamline MI diagnosis, lessen the burden on clinicians, and notably influence the feasibility of a clinical MI detection device.
Data Availability Statement
The datasets used and/or analyzed during in current study are available from the public Physionet PTB dataset [61, 62] which can be accessed at the website: https://www.physionet.org/content/ptbdb/1.0.0/.
References
Liu B, Liu J, Wang G, Huang K, Li F, Zheng Y, Zhou F (2015) A novel electrocardiogram parameterization algorithm and its application in myocardial infarction detection. Comput Biol Med 61:178–184
Strodthoff N, Strodthoff C (2019) Detecting and interpreting myocardial infarction using fully convolutional neural networks. Physiol Measure 40(1):015001
Tripathy RK, Dandapat S (2016) Detection of cardiac abnormalities from multilead ECG using multiscale phase alternation features. J Med Syst 40(6):143
Kloner RA, Darsee JR, DeBoer LW, Carlson N (1981) Early pathologic detection of acute myocardial infarction. Arch Pathol Lab Med 105(8):403–406
Visser CA, Lie KI, Kan G, Meltzer R, Durrer D (1981) Detection and quantification of acute, isolated myocardial infarction by two dimensional echocardiography. Am J Cardiol 47(5):1020–1025
Gibler WB, Lewis LM, Erb RE, Makens PK, Kaplan BC, Vaughn RH, Campbell WB (1990) Early detection of acute myocardial infarction in patients presenting with chest pain and nondiagnostic ECGs: serial CK-MB sampling in the emergency department. Ann Emergency Med 19(12):1359–1366
Ohman EM, Casey C, Bengtso J, Pryor D, Tormey W, Horgan JH (1990) Early detection of acute myocardial infarction: additional diagnostic information from serum concentrations of myoglobin in patients without ST elevation. Heart 63(6):335–338
Shah CP, Thakur RK, Reisdorff EJ, Lane E, Aufderheide TP, Hayes OW (1998) QT dispersion may be a useful adjunct for detection of myocardial infarction in the chest pain center. Am Heart J 136(3):496–498
Huber AM, Schoenberg SO, Hayes C, Spannagl B, Engelmann MG, Franz WM, Reiser MF (2005) Phase-sensitive inversion-recovery MR imaging in the detection of myocardial infarction. Radiology 237(3):854–860
Li R, Zhao X, Gong Y, Zhang J, Dong R, Xia L (2021) A new method for detecting myocardial ischemia based on ECG T-wave area curve (TWAC). Frontiers in Physiology 12
Acharya UR, Fujita H, Adam M, Lih OS, Sudarshan VK, Hong TJ, San TR (2017) Automated characterization and classification of coronary artery disease and myocardial infarction by decomposition of ECG signals: A comparative study. Inform Sci 377:17–29
Sharma H, Sharma KK (2018) ECG-derived respiration using Hermite expansion. Bio Signal Process Control 39:312–326
Geddes J, Mehlsen J, Olufsen MS (2020) Characterization of blood pressure and heart rate oscillations of POTS patients via uniform phase empirical mode decomposition. IEEE Trans Biomed Eng 67(11):3016–3025
Hasan NI, Bhattacharjee A (2019) Deep learning approach to cardiovascular disease classification employing modified ECG signal from empirical mode decomposition. Biomed Signal Process Control 52:128–140
Khan S, Pachori RB (2021) Automated detection of posterior myocardial infarction from vectorcardiogram signals using Fourier bessel series expansion based empirical wavelet transform. IEEE Sens Lett 5(5):7001604
Zeng W, Yuan J, Yuan C, Wang Q, Liu F, Wang Y (2020) Classification of myocardial infarction based on hybrid feature extraction and artificial intelligence tools by adopting tunable-Q wavelet transform (TQWT), variational mode decomposition (VMD) and neural networks. Artificial Intell Med 106:101848
Sadhukhan D, Pal S, Mitra M (2018) Automated identification of myocardial infarction using harmonic phase distribution pattern of ECG data. IEEE Trans Instrumentation Measure 67(10):2303–2313
Butun E, Yildirim O, Talo M, Tan RS, Acharya UR (2020) 1D-CADCapsNet: One dimensional deep capsule networks for coronary artery disease detection using ECG signals. Physica Medica 70:39–48
Sudarshan VK, Acharya UR, Oh SL, Adam M, Tan JH, Chua CK, San Tan R (2017) Automated diagnosis of congestive heart failure using dual tree complex wavelet transform and statistical features extracted from 2 s of ECG signals. Comput Biol Med 83:48–58
Dohare AK, Kumar V, Kumar R (2018) Detection of myocardial infarction in 12 lead ECG using support vector machine. Appl Soft Comput 64:138–147
Singh V, Gupta A, Sohal JS, Singh A (2019) A unified non-linear approach based on recurrence quantification analysis and approximate entropy: application to the classification of heart rate variability of age-stratified subjects. Med Biol Eng Comput 57(3):741–755
Yang X, Wang Z, He A, Wang J (2020) Identification of healthy and pathological heartbeat dynamics based on ECG-waveform using multifractal spectrum. Physica A 559:125021
Han C, Shi L (2019) Automated interpretable detection of myocardial infarction fusing energy entropy and morphological features. Comput Methods Programs Biomed 175:9–23
Mazaheri V, Khodadadi H (2020) Heart arrhythmia diagnosis based on the combination of morphological, frequency and nonlinear features of ECG signals and metaheuristic feature selection algorithm. Expert Syst Appl 161:113697
Adam M, Oh SL, Sudarshan VK, Koh JE, Hagiwara Y, Tan JH, Acharya UR (2018) Automated characterization of cardiovascular diseases using relative wavelet nonlinear features extracted from ECG signals. Comput Methods Programs Biomed 161:133–143
Liu J, Zhang C, Zhu Y, Ristaniemi T, Parviainen T, Cong F (2020) Automated detection and localization system of myocardial infarction in single-beat ECG using Dual-Q TQWT and wavelet packet tensor decomposition. Comput Methods Programs Biomed 184:105120
Sun Q, Xu Z, Liang C, Zhang F, Li J, Liu R, Wang C (2023) A dynamic learning-based ECG feature extraction method for myocardial infarction detection. Physiol Measure 43(12):124005
Chauhan C, Tripathy RK, Agrawal M (2023) Patient specific higher order tensor based approach for the detection and localization of myocardial infarction using 12-lead ECG. Biomed Signal Process Control 83:104701
Doudesis D, Lee KK, Boeddinghaus J, Bularga A, Ferry AV, Tuck C, Mills NL (2023) Machine learning for diagnosis of myocardial infarction using cardiac troponin concentrations. Nat Med 29:1201–1210
Oliveira M, Seringa J, Pinto FJ, Henriques R, Magalhaes T (2023) Machine learning prediction of mortality in Acute Myocardial Infarction. BMC Med Inform Decision Making 23(1):1–16
Deng M, Huang X, Liang Z, Lin W, Mo B, Liang D, Chen J (2023) Classification of cardiac electrical signals between patients with myocardial infarction and normal subjects by using nonlinear dynamics features and different classification models. Biomed Signal Process Control 79:104105
Arif M, Malagore IA, Afsar FA (2012) Detection and localization of myocardial infarction using k-nearest neighbor classifier. J Med Syst 36(1):279–289
Lagerholm M, Peterson C, Braccini G, Edenbrandt L, Sornmo L (2000) Clustering ECG complexes using Hermite functions and self-organizing maps. IEEE Trans Biomed Eng 47(7):838–848
Zhang J, Lin F, Xiong P, Du H, Zhang H, Liu M, Liu X (2019) Automated detection and localization of myocardial infarction with staked sparse autoencoder and treebagger. IEEE Access 7:70634–70642
Shouval R, Hadanny A, Shlomo N, Iakobishvili Z, Unger R, Zahger D, Beigel R (2017) Machine learning for prediction of 30-day mortality after ST elevation myocardial infraction: An acute coronary syndrome Israeli survey data mining study. Int J Cardiol 246:7–13
Yang P, Wang D, Zhao WB, Fu LH, Du JL, Su H (2021) Ensemble of kernel extreme learning machine based random forest classifiers for automatic heartbeat classification. Biomed Signal Process Control 63:102138
Costa CM, Silva IS, de Sousa RD, Hortegal RA, Regis CDM (2018) The association between reconstructed phase space and Artificial Neural Networks for vectorcardiographic recognition of myocardial infarction. J Electrocardiol 51(3):443–449
Acharya UR, Fujita H, Lih OS, Adam M, Tan JH, Chua CK (2017) Automated detection of coronary artery disease using different durations of ECG segments with convolutional neural network. Knowl-Based Syst 132:62–71
Lih OS, Jahmunah V, San TR, Ciaccio EJ, Yamakawa T, Tanabe M, Acharya UR (2020) Comprehensive electrocardiographic diagnosis based on deep learning. Artificial Intell Med 103:101789
Hernandez-Matamoros A, Fujita H, Meana HMP (2020) Recognition of heartbeat categories applying a novel preprocessing scheme and neural networks. SoMeT, Frontiers in Artificial Intelligence and Applications, pp. 162-172
Madhavi KR, Kora P, Reddy LV, Avanija J, Soujanya KLS, Telagarapu P (2022) Cardiac arrhythmia detection using dual-tree wavelet transform and convolutional neural network. Soft Comput 26(7):3561–3571
Hernandez AA, Bonizzi P, Peeters R, Karel J (2023) Continuous monitoring of acute myocardial infarction with a 3-Lead ECG system. Biomed Signal Process Control 79:104041
Rai HM, Chatterjee K (2022) Hybrid CNN-LSTM deep learning model and ensemble technique for automatic detection of myocardial infarction using big ECG data. Appl Intell 52(5):5366–5384
Li W, Tang YM, Yu KM, To S (2022) SLC-GAN: An automated myocardial infarction detection model based on generative adversarial networks and convolutional neural networks with single-lead electrocardiogram synthesis. Inform Sci 589:738–750
Tripathy RK, Bhattacharyya A, Pachori RB (2019) A novel approach for detection of myocardial infarction from ECG signals of multiple electrodes. IEEE Sens J 19(12):4509–4517
Chuang YH, Huang CL, Chang WW, Chien JT (2020) Automatic classification of myocardial infarction using spline representation of single-lead derived vectorcardiography. Sensors 20(24):7246
Acharya UR, Fujita H, Oh SL, Hagiwara Y, Tan JH, Adam M (2017) Application of deep convolutional neural network for automated detection of myocardial infarction using ECG signals. Inform Sci 415:190–198
Hong S, Zhou Y, Shang J, Xiao C, Sun J (2020) Opportunities and challenges of deep learning methods for electrocardiogram data: a systematic review. Comput Biol Med 122:103801
Cheffer A, Savi MA, Pereira TL, de Paula AS (2021) Heart rhythm analysis using a nonlinear dynamics perspective. Appl Math Modell 96:152–176
Fojt O, Holcik J (1998) Applying nonlinear dynamics to ECG signal processing. IEEE Eng Med Biol Mag 17(2):96–101
Owis MI, Abou-Zied AH, Youssef AB, Kadah YM (2002) Study of features based on nonlinear dynamical modeling in ECG arrhythmia detection and classification. IEEE Trans Biomed Eng 49(7):733–736
Nayak SK, Bit A, Dey A, Mohapatra B, Pal K (2018) A review on the nonlinear dynamical system analysis of electrocardiogram signal. Journal of Healthcare Engineering 2018
Al-Fahoum AS, Qasaimeh AM (2013) A practical reconstructed phase space approach for ECG arrhythmias classification. J Med Eng Technol 37(7):401–408
Wang Z, Ning X, Zhang Y, Du G (2000) Nonlinear dynamic characteristics analysis of synchronous 12-lead ECG signals. IEEE Eng Med Biol Mag 19(5):110–115
Abdalla FY, Wu L, Ullah H, Ren G, Noor A, Zhao Y (2019) ECG arrhythmia classification using artificial intelligence and nonlinear and nonstationary decomposition. Signal Image Video Process 13(7):1283–1291
Zeeman EC (1972) Differential equations for the heartbeat and nerve impulse. University of Warwick, Coventry, UK, Mathematics Institute
Zeeman EC (1972). Differential equations for the heartbeat and nerve impulse. In: Waddington, C.H. (Ed.), Towards a Theoretical Biology, vol. 4. Edinburgh University Press
Suckley R, Biktashev VN (2003) Comparison of asymptotics of heart and nerve excitability. Phys Rev E 68(1):011902
Jafarnia-Dabanloo N, McLernon DC, Zhang H, Ayatollahi A, Johari-Majd V (2007) A modified Zeeman model for producing HRV signals and its application to ECG signal generation. J Theoretical Biol 244(2):180–189
Das S, Maharatna K (2013) Fractional dynamical model for the generation of ECG like signals from filtered coupled Van-der Pol oscillators. Comput Methods Programs Biomed 112(3):490–507
Bousseljot R, Kreiseler D, Schnabel A. Nutzung der EKG-Signaldatenbank CARDIODAT der PTB über das Internet.Biomedizinische Technik 40(s1): 317-318
Goldberger AL, Amaral LA, Glass L, Hausdorff JM, Ivanov PC, Mark RG, Stanley HE. PhysioBank, PhysioToolkit, and PhysioNet: components of a new research resource for complex physiologic signals. Circulation 101(23): e215-e220
Kumar M, Pachori RB, Acharya UR (2017) Automated diagnosis of myocardial infarction ECG signals using sample entropy in flexible analytic wavelet transform framework. Entropy 19(9):488
Oh SL, Ng EY, San Tan R, Acharya UR (2018) Automated diagnosis of arrhythmia using combination of CNN and LSTM techniques with variable length heart beats. Comput Biol Med 102:278–287
Sharma LD, Sunkaria RK (2018) Inferior myocardial infarction detection using stationary wavelet transform and machine learning approach. Signal Image Video Process 12(2):199–206
Kligfield P, Gettes LS, Bailey JJ, Childers R, Deal BJ, Hancock EW, Wagner GS (2007) Recommendations for the standardization and interpretation of the electrocardiogram: part I: the electrocardiogram and its technology a scientific statement from the American Heart Association Electrocardiography and Arrhythmias Committee, Council on Clinical Cardiology; the American College of Cardiology Foundation; and the Heart Rhythm Society endorsed by the International Society for Computerized Electrocardiology. J Am College Cardiol 49(10):1109–1127
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. J Artificial Intell Res 16:321–357
Feng W, Dauphin G, Huang W, Quan Y, Bao W, Wu M, Li Q (2019) Dynamic synthetic minority over-sampling tTechnique-based rotation forest for the classification of imbalanced hyperspectral data. IEEE J Selected Topics Appl Earth Observations Remote Sens 12(7):2159–2169
Wang Q, Zhou X, Wang C, Liu Z, Huang J, Zhou Y, Cheng JZ (2019) WGAN-based synthetic minority over-sampling technique: improving semantic fine-grained classification for lung nodules in CT images. IEEE Access 7:18450–18463
Rivera WA, Xanthopoulos P (2016) A priori synthetic over-sampling methods for increasing classification sensitivity in imbalanced data sets. Expert Syst Appl 66:124–135
Keunen RW, Pijlman HG, Visee HF, Vliegen JH, Tavy DL, Stam KJ (1994) Dynamical chaos determines the variability of transcranial Doppler signals. Neurol Res 16(5):353–358
Smidtaite R, Navickas Z, Venskaityte E (2010) ECG research using elements of matrix analysis and phase planes. Elektronika ir Elektrotechnika 103(7):83–86
Beyramienanlou H, Lotfivand N (2017) Shannon’s energy based algorithm in ECG signal processing. Computational and Mathematical Methods in Medicine 2017, Article ID 8081361
Huang NE, Shen Z, Long SR, Wu MC, Shih HH, Zheng Q, Liu HH (1998) The empirical mode decomposition and Hilbert spectrum for nonlinear and non-stationary time series analysis. Proceedings of the Royal Society of London A: Mathematical, Physical and Engineering Sciences. The Royal Society 454(1971): 903-995
Thirumalaisamy MR, Ansell PJ (2018) Fast and adaptive empirical mode decomposition for multidimensional, multivariate signals. IEEE Signal Process Lett 25(10):1550–1554
Wang C, Hill DJ (2006) Learning from neural control. IEEE Trans Neural Netw 17(1):130–146
Wang C, Hill DJ (2007) Deterministic learning and rapid dynamical pattern recognition. IEEE Trans Neural Netw 18(3):617–630
Wang C, Hill DJ (2009) Deterministic Learning Theory for Identification. CRC Press, Boca Raton, FL, Recognition and Control
Zhang J, Liu M, Xiong P, Du H, Zhang H, Lin F, Liu X (2021) A multi-dimensional association information analysis approach to automated detection and localization of myocardial infarction. Eng Appl Artificial Intell 97:104092
Jafarian K, Vahdat V, Salehi S, Mobin M (2020) Automating detection and localization of myocardial infarction using shallow and end-to-end deep neural networks. Appl Soft Comput 93:106383
Jian JZ, Ger TR, Lai HH, Ku CM, Chen CA, Abu PAR, Chen SL (2021) Detection of myocardial infarction using ECG and multi-scale feature concatenate. Sensors 21(5): 1906
Hammad M, Alkinani MH, Gupta BB, El-Latif A, Ahmed A (2022) Myocardial infarction detection based on deep neural network on imbalanced data. Multimedia Syst 28(4):1373–1385
Wagner P, Strodthoff N, Bousseljot RD, Kreiseler D, Lunze FI, Samek W, Schaeffter T (2020) PTB-XL, a large publicly available electrocardiography dataset. Sci Data 7(1):154
Acknowledgements
This work was supported by the Natural Science Foundation of Fujian Province (Grant no. 2022J011146).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zeng, W., Shan, L., Yuan, C. et al. Detection of myocardial infarction using Shannon energy envelope, FA-MVEMD and deterministic learning. Complex Intell. Syst. (2024). https://doi.org/10.1007/s40747-024-01419-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40747-024-01419-x