Abstract
The existing DUS-multivariate inverse Weibull distribution under classical statistics can be applied when all observations in the data are imprecise. In this paper, we introduce DUS-neutrosophic multivariate inverse Weibull distribution that can be used when the observations in the data are imprecise or in intervals. We derive some statistical properties and functions of DUS-neutrosophic multivariate inverse Weibull distribution. We also discuss the maximum likelihood estimation method for estimating the parameters. Monte-Carlo simulation study is performed to study the behavior of maximum likelihood estimates. We compare the efficiency of the proposed DUS-neutrosophic multivariate inverse Weibull distribution with the existing distributions under classical statistics. From the comparison, it is found that the proposed DUS-neutrosophic multivariate inverse Weibull distribution provides smaller values of Akaike’s information criteria and Bayesian information criteria than the existing distributions under classical statistics. The proposed study can be extended for other statistical distributions as future research.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Multivariate distributions are very important in many applications in real life also it is important in studying multivariate analysis, regression analysis, and so on. Many methods are used to construct the bivariate and multivariate distributions such as various copula functions see [2,3,4, 14]. In this study, we are interested in multivariate inverse Weibull distribution. The main aim of choosing this distribution is the inverse Weibull distribution is a lifetime probability distribution that can be used in reliability, engineering, and biological studies for more details see [16, 24]. Inverse Weibull distribution is used also as a stress-strength model see [11].
Definition 1
If a random variable \(Y\) has Weibull distribution, then the random variable \(X=\frac{1}{Y}\) follows the inverse Weibull distribution denoted as \(\mathrm{IWD}(\alpha ,\beta )\). Then the probability density function (pdf) and cumulative distribution function (CDF) of \(X\) are defined as follow, respectively.
where, \(x, \alpha , \beta >0\).
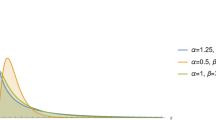
Copula function is a more suitable approach to get bivariate and multivariate distributions for more details see [18] (Fig. 1).
Definition 2
Let a random vector \(({X}_{1}\dots {X}_{d})\) and the marginal CDFs denoted by \({F}_{i}\left(x\right)=P[{X}_{i}<x]\) for \(i=1\dots d.\) Then using probability integral transformation (PIT) for each component, the distribution of the random vector \(\left({U}_{1}\dots {U}_{d}\right)=({F}_{1}\left({X}_{1}\right)\dots {F}_{d}\left({X}_{d}\right))\) belongs to the (\(\mathrm{unif}{(\mathrm{0,1})}^{d})\) family distribution and the copula related to \(({X}_{1}\dots {X}_{d})\) is defined as the joint CDF of \(\left({U}_{1}\dots {U}_{d}\right)\),i.e.
With \(\left({u}_{1}\dots {u}_{d}\right)\in {[\mathrm{0,1}]}^{d}\).
Definition 3
\(\mathrm{C}:{[\mathrm{0,1}]}^{d}\to [\mathrm{0,1}]\) is a d-dimensional copula if it’s a CDF with \(C\left({u}_{1}\dots .{u}_{i-1},0,{u}_{i+1}\dots {u}_{d}\right)=0\), \(C\left(1\dots u\dots .1\right)=u,\) with \(\left({u}_{1},\dots ,{u}_{d}\right)\in {[\mathrm{0,1}]}^{d}\) and \(u\in \left[\mathrm{0,1}\right].\)
In the context of Multivariate inverse Weibull which introduced by Al-Hussaini and Ateya [3] and denoted as \(\mathrm{MIWD}({\alpha }_{i},{\beta }_{i},\gamma )\) using copula approach as follows.
Definition 4
Let \({\varvec{X}}=({X}_{1}\dots {X}_{m})\) is a random vector of random variables such that \({X}_{i}\sim \mathrm{IWD}({\alpha }_{i},{\beta }_{i})\),\(i=1\dots m\). Then the CDF and pdf of \({\varvec{X}}\) are defined as
where \(\gamma >0, \boldsymbol{\alpha }=({\alpha }_{1}\dots {\mathrm{\alpha }}_{{\varvec{m}}})\) and \({\varvec{\beta}}=({\beta }_{1}\dots {\upbeta }_{{\varvec{m}}})\) (Fig. 2).
In recent years, neutrosophic statistics is appear and are defined as an extension of classical statistics and one deal with set values instead of crisp values. Ashbacher [5] introduced fuzzy logic and neutrosophy, for more details on neutrosophic statistics, see [25, 26, 28]. Many authors interested in neutrosophic probability distribution and are shown that it is better than the classical statistics in real life problems such as [25] introduced the neutrosophic Weibull distribution, [22, 23] introduced neutrosophic beta distribution, Patro and Smarandache [19] introduced neutrosophic normal distribution and neutrosophic binomial distribution. These new distributions allowed us to solve more problems depending on indeterminacy which is ignored in classical statistics. Some authors constructed the new distributions using DUS-transformation which is introduced by [15] and used it to construct a new distribution bounded within [0,1].
Definition 5
Let \(f\left(x\right)\), \(F(x)\) and \(h(x)\) denote, respectively, probability density function, the cumulative distribution function, and the hazard rate function of baseline distribution. Then the DUS-family are given by
where \(x\in D\) and \(D\) is a domain of the baseline distribution.
Many authors used the DUS-transformation to construct new distributions such as [15] introduced DUS-transformation with an exponential distribution, Deepthi and Chacko [6] introduced DUS-transformation with the Lomax distribution and Karakaya [12] introduced DUS-transformation with the Kumaraswamy distribution. Nayana et al. [17] introduced DUS-transformation with neutrosophic Weibull distribution. More details about the applications of neutrosophic statistics can be seen [1, 7, 10, 13, 20,21,22,23]. The idea for improving the quality aspect can be seen in Ghosh et al. [9].
Since multivariate analysis is one of the most useful methods to determine relationships and analyze patterns among large sets of data. It is particularly effective in minimizing bias if a structured study design is employed. However, the complexity of the technique makes it a less sought-out model for novice research enthusiasts. Therefore, although the process of designing the study and interpretation of results is a tedious one, the techniques stand out in finding the relationships in complex situations. Since the multivariate analysis depends on the multivariate distribution, then we need more multivariate distributions also for indeterminacies in real data, then we need to construct neutrosophic multivariate distributions. A rich literature on DUS-transformation is available under classical statistics. The existing distributions cannot be applied when the data have imprecise observations. To overcome this issue, we will introduce DUS-MIWD which can be applied when the data has imprecise observations. By exploring the literature and according to the best of our knowledge; there is no work on DUS-MIWD under neutrosophic statistics. To fill this gap, we will propose DUS-MIWD under neutrosophic statistics. Then our aim in this study is to extend the use of DUS-transformation to construct new distribution called DUS-neutrosophic multivariate inverse Weibull distribution and denoted by DUS-NMIWD. This paper is organized as follows: the pdf, CDF, and hazard function of our new distribution are introduced, some statistical properties of our new distribution are obtained, an estimation of distribution parameters is obtained, a simulation study was performed to discuss the distribution parameters is introduced, the real example is introduced .
DUS-neutrosophic multivariate inverse Weibull
Suppose that \({I}_{N}\in ({I}_{L},{I}_{U})\) is an indeterminacy interval \(N\) is neutrosophic statistical number and let \({X}_{smN}={X}_{smL}+{X}_{smU}{I}_{N}\), be a random vector of random variables follow neutrosophic multivariate inverse Weibull \({X}_{smN}\) be a random vector with m-variables and S-observations. If \({X}_{N}\in [{n}_{L},{n}_{U}]\) are observations of neutrosophic from neutrosophic variable \({P}_{N}\in [{P}_{L},{P}_{U}]\), then
The form of neutosophic \({X}_{N}={X}_{L}+{X}_{N} {I}_{N}\), \({I}_{N}\in [{I}_{L},{I}_{U}]\) and in multivariate case \({X}_{mN}={X}_{mL}+{X}_{mN} {I}_{N}\). Therefore, we consider neutrosophic multivariate inverse Weibull distribution of a random vector \({{\varvec{X}}}_{{\varvec{N}}}=({X}_{1N}\dots {X}_{mN})\) with neutrosophic joint probability distribution function (njpdf) as
The corresponding neutrosophic joint cumulative distribution (njCDF) is
Using Eqs. (8) and (9) in Dus-transformation then we get DUS-neutrosophic multivariate inverse Weibull distribution (DUS-NMIWD) with njpdf, njCDF, and NJhf as follow, respectively (Figs. 3, 4, 5).
and,
Statistical properties of DUS-NMIWD
In this section, we obtained some important statistical properties of DUS-NMIWD, when \(m=2\), is called DUS-neutrosophic bivariate inverse Weibull distribution and is denoted by DUS-NBIWD. The neutrosophic joint probability density function, the neutrosophic joint cumulative distribution function, the neutrosophic joint hazard function, the neutrosophic marginal distribution, the neutrosophic product moments, the neutrosophic moment generating function, the neutrosophic conditional distribution and neutrosophic reliability function.
The neutrosophic joint probability density function
where,\({{ \alpha }_{1},\alpha }_{2},{\beta }_{1},{\beta }_{2},\gamma >0\), \({x}_{1N},{x}_{2N}>0,{x}_{1N},{x}_{2N}\in \left[{X}_{mL},{X}_{mU}\right], m=\mathrm{1,2}\) and \({I}_{k}\in \left[{I}_{L},{I}_{U}\right].\)
The neutrosophic joint cumulative distribution function
The neutrosophic joint hazard function
The neutrosophic marginal distribution
The neutrosophic marginal distributions of \({X}_{1N}\) and \({X}_{2N}\) are getting as follows
where, \(i,j=\mathrm{1,2}\) and \(i\ne j\)
Then the neutrosophic marginal distributions of \({X}_{1N}\) and \({X}_{2N}\) are
And,
Neutrosophic product moments
If the random vector \({(x}_{1N},{x}_{2N})\) is distributed as DUS-NBIWD, then the rth and sth moments about the origin is given by
Neutrosophic moment generating function
If the random vector \({(x}_{1N},{x}_{2N})\) is distributed as DUS-NBIWD, then the moment generating function is defined by
Neutrosophic conditional distribution
The conditional probability distribution of \({X}_{1N}\) given \({X}_{2N}\) is given as follows
Also, we can get the conditional probability distribution of \({X}_{2N}\) given \({X}_{1N}\) is given as follows
Neutrosophic reliability function
If the random vector \({(x}_{1N},{x}_{2N})\) is distributed as DUS-NBIWD, then the reliability function is denoted by \({R}_{\mathrm{DUS}}\left({x}_{1N},{x}_{2N};{\alpha }_{1},{\alpha }_{2},{\beta }_{1},{\beta }_{2},\gamma \right)\) and given by
Maximum likelihood estimation method
To estimate the parameters of our new distribution we use the maximum likelihood estimation method which introduced by Elaal and Jarwan [8] for all joint models. Now, the maximum likelihood estimates of the vector \(\Theta =({\alpha }_{1},{\alpha }_{2},{\beta }_{1},{\beta }_{2},\gamma )\) is given as follows,
-
1.
Construct the likelihood function as
$$L\left({X}_{1N},{X}_{2N}\left|\Theta \right.\right)=\prod_{i=1}^{n}{f}_{\mathrm{DUS}}\left({x}_{1Ni},{x}_{2Ni};{\alpha }_{1},{\alpha }_{2},{\beta }_{1},{\beta }_{2},\gamma \right)$$ -
2.
Take Log for likelihood function as
$$\begin{aligned}&\mathrm{Log} L({X}_{1N},{X}_{2N}|\Theta )\\&=\sum_{i=1}^{n}\mathrm{Log}([({x}_{1Ni},{x}_{2Ni};{\alpha }_{1},{\alpha }_{2},{\beta }_{1},{\beta }_{2},\gamma ))]\\&=(n+1)(\mathrm{Log}[(1+{I}_{k})])+n(\mathrm{Log}[{\alpha }_{1}]\\&\quad +\mathrm{Log}[{\beta }_{1}]+\mathrm{Log}[{\alpha }_{2}]+\mathrm{Log}[{\beta }_{2}]+\mathrm{Log}[\Gamma [2+\gamma ]])\\&\quad -n (\mathrm{Log}[e-1]+\mathrm{Log}[\gamma ]+\mathrm{Log}[\Gamma [\gamma ]])\\&\quad -(1+{\alpha }_{1})\sum_{i=1}^{n}{x}_{1Ni}-(1+{\alpha }_{2})\sum_{i=1}^{n}{x}_{2Ni}\\&\quad -\gamma \sum_{i=1}^{n}\left({e}^{\frac{{\beta }_{1}{x}_{1Ni}^{-{\alpha }_{1}}}{\gamma }}+{e}^{\frac{{\beta }_{2}{x}_{2Ni}^{-{\alpha }_{2}}}{\gamma }}-1\right)\\&\quad +\sum_{i=1}^{n}\frac{{\beta }_{1}{x}_{1Ni}^{-{\alpha }_{1}}}{\gamma }+\sum_{i=1}^{n}\frac{{\beta }_{2}{x}_{2Ni}^{-{\alpha }_{2}}}{\gamma }\\&\quad -(2+\gamma )\sum_{i=1}^{n}\mathrm{Log}\left[{e}^{\frac{{\beta }_{1}{x}_{1N}^{-{\alpha }_{1}}}{\gamma }}+{e}^{\frac{{\beta }_{2}{x}_{2N}^{-{\alpha }_{2}}}{\gamma }}-1\right]\end{aligned}$$ -
3.
Get the normal equations for \({(\alpha }_{1},{\alpha }_{2},{\beta }_{1},{\beta }_{2},\gamma \))
$$\begin{aligned}\frac{n}{{\widehat{\alpha }}_{1}}&-\sum_{i=1}^{n}{x}_{1Ni}+ \widehat{\gamma }\sum_{i=1}^{n}\frac{{e}^{\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}}{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}\mathrm{Log}\left[{x}_{1Ni}\right]}{\widehat{\gamma }}\\&-\sum_{i=1}^{n}\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}+\left(2+\widehat{\gamma }\right)\\&\times\sum_{i=1}^{n}\frac{{e}^{\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}}{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}\mathrm{Log}\left[{x}_{1Ni}\right]}{\left({e}^{\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}}+{e}^{\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{\widehat{\gamma }}}-1\right)\widehat{\gamma }}=0\end{aligned}$$$$\begin{aligned}\frac{n}{{\widehat{\alpha }}_{2}}&-\sum_{i=1}^{n}{x}_{2Ni}+ \widehat{\gamma }\sum_{i=1}^{n}\frac{{e}^{\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{\widehat{\gamma }}}{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}\mathrm{Log}\left[{x}_{2Ni}\right]}{\widehat{\gamma }}\\&-\sum_{i=1}^{n}\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{\widehat{\gamma }}+\left(2+\widehat{\gamma }\right)\\&\times\sum_{i=1}^{n}\frac{{e}^{\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{\widehat{\gamma }}}{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}\mathrm{Log}\left[{x}_{2Ni}\right]}{\left({e}^{\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}}+{e}^{\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{\widehat{\gamma }}}-1\right)\widehat{\gamma }}=0\end{aligned}$$$$\begin{aligned}\frac{n}{{\widehat{\beta }}_{1}}&-\sum_{i=1}^{n}{e}^{\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}}{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}\\&+\sum_{i=1}^{n}\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}}{\widehat{\gamma }}-\left(2+\widehat{\gamma }\right)\\&\times\sum_{i=1}^{n}\frac{{e}^{\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}}{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}}{\left({e}^{\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}}+{e}^{\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{\widehat{\gamma }}}-1\right)\widehat{\gamma }}=0\end{aligned}$$$$\begin{aligned}\frac{n}{{\widehat{\beta }}_{2}}&-\sum_{i=1}^{n}{e}^{\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{\widehat{\gamma }}}{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}\\&+\sum_{i=1}^{n}\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}}{\widehat{\gamma }}-\left(2+\widehat{\gamma }\right)\\&\times\sum_{i=1}^{n}\frac{{e}^{\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}}{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}}{\left({e}^{\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}}+{e}^{\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{\widehat{\gamma }}}-1\right)\widehat{\gamma }}=0\end{aligned}$$$$\begin{aligned}&n\mathrm{PolyGamma}\left[\mathrm{0,2}+\widehat{\gamma }\right]-n \mathrm{PolyGamma}\left[0,\widehat{\gamma }\right]\\&-\frac{{n}}{\widehat{\gamma }}+\widehat{\gamma }\sum_{i=1}^{n}\left(\frac{{e}^{\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}}{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{{\widehat{\gamma }}^{2}}+\frac{{e}^{\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{\widehat{\gamma }}}{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{{\widehat{\gamma }}^{2}}\right)\\&+\left(2+\widehat{\gamma }\right) \sum_{i=1}^{n}\frac{\left(\frac{{e}^{\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}}{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{{\widehat{\gamma }}^{2}}+\frac{{e}^{\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{\widehat{\gamma }}}{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{{\widehat{\gamma }}^{2}}\right)}{\left({e}^{\frac{{{x}_{1Ni}}^{-{\widehat{\alpha }}_{1}}{\widehat{\beta }}_{1}}{\widehat{\gamma }}}+{e}^{\frac{{{x}_{2Ni}}^{-{\widehat{\alpha }}_{2}}{\widehat{\beta }}_{2}}{\widehat{\gamma }}}-1\right)}=0\end{aligned}$$
We can not solve normal equations analytically, so we use a numerical method to get the maximum likelihood estimates for our distribution parameters.
Monte-Carlo simulation study
Monte-Carlo simulation study is used to show the performance of our new distribution when we compare the maximum likelihood estimates of our new distribution DUS-NMIWD with their corresponding maximum likelihood estimates of classical version DUS-MIWD according to two statistical criteria bias and MSE. To perform the Monte-Carlo simulation study we use the following steps
-
1.
Assume the different initial values of our distribution parameters \(\left({\alpha }_{1},{\alpha }_{2},{\beta }_{1},{\beta }_{2},\gamma \right)=\left(\mathrm{0.5,0.6,1.5,2.5,0.3}\right), \left(1.5,2.5,0.5,0.8,3\right), \left(1.5,2.5,2,\mathrm{3,0.5}\right).\)
-
2.
For our new distribution we use two indeterminacy measures \({I}_{N}=(\mathrm{0.2,0.5})\) and\({I}_{N}=(\mathrm{0.6,0.8})\). Note when \({I}_{N}=0\), then we get the classical DUS-IWD.
-
3.
Generate different sample sizes \(n=50, 100, 200, 500\). Using the different initial values of our distribution parameters which assumed in step one.
-
4.
Find the maximum likelihood estimates of the parameters which denoted by \({(\widehat{\alpha }}_{1},{\widehat{\alpha }}_{2},{\widehat{\beta }}_{1},{\widehat{\beta }}_{2},\widehat{\gamma })\).
-
5.
Calculate bias and mean square error.
Tables 1 and 2 show the results of the simulation study for our new distribution. Table 3 shows the result of the simulation study for the classical version of our distribution. From Tables 1, 2, 3 we get the bias and Mean Square Error (MSE) for our new distribution DUS-NMIWD is smaller than their corresponding for the classical version DUS-MIWD. So, we can decide whether the new distribution is better than the classical version.
Comparative studies using real data
To show the performance of our new distribution DUS-NMIWD (bivariate case), we compare it and other distributions i.e., DUS-MIWD, NMIWD and MIWD (bivariate case) on real data which represent the data of the quality control division for quality characteristics of glass production see [27]. There are two quality characteristics such as cutter line \({X}_{1N}\) and Edge distortion \({X}_{2N}\). The target of the cutter is 115 mm, and the Edge distortion is 40 mm. The data is shown in Table 4 as follows.
Figure 6 shows the architectural diagram of the proposed algorithm in this section. In this section, we use two statistical criteria to test the performance of our new distribution. AIC (Akaike’s Information Criteria) and BIC (Bayesian Information Criteria), where,
where, \(\underline{\Theta }\): the vector of distribution parameters, \(L\left(\underline{\Theta }\right)\): the likelihood function, \( k\): the number of estimates, n: the data size. The small value of AIC and BIC means good-fit distribution. Table 5 shows the result of the comparison between our new distribution and other distributions under classical statistics. From Table 5, we get the AIC = (1175.6, 1341.29) and BIC = (1175.81, 1342.62) so it is smaller than other distributions therefore, the DUS-NMIW is the best for data than the other distributions under classical statistics.
Limitations
As mentioned earlier that the neutrosophic statistics is an extension of classical statistics and applied when the observations in the data are imprecise. The proposed DUS-NMIWD has advantages over the existing DUS-MIWD under classical statistics in terms of AIC and BIC. From Table 5, it is clear that the proposed DUS-NMIWD provides smaller values of AIC and BIC as compared to the existing distributions under classical statistics. Therefore, for the interval data, the use of the proposed DUS-NMIWD can be recommended over the existing distributions under classical statistics. The proposed DUS-NMIWD under neutrosophic statistics has some limitations such as it cannot be applied when the multivariate data has no imprecise observations. In addition, the proposed DUS-NMIWD can be applied only when the quality of characteristics follows the normal distribution.
Conclusions
The main aim of this paper was to introduce the new distribution is called DUS-neutrosophic multivariate inverse Weibull distribution. The proposed DUS-NMIWD was the generalization of DUS-MIWD under classical statistics. The proposed DUS-NMIWD reduces to the existing DUS-MIWD when no indeterminate observations are found in the data. We derived some properties of the proposed distribution. The simulation study and the analysis of real examples showed that the proposed DUS-NMIWD performs better than the existing DUS-MIWD under uncertain environments. The proposed DUS-NMIWD can be applied to reliability, and survival analysis, in many engineering fields and medicine when the data have imprecise observations. The proposed DUS-NMIWD can be extended for non-normal distributions as future research. The proposed distribution can be used in a designing control chart for future research.
Data availability
The data is given in the paper.
References
Alhabib R, Ranna MM, Farah H, Salama A (2018) Some neutrosophic probability distributions. Neutrosoph Sets Syst 22:30–38
Al-Hussaini EK, Ateya SF (2005) Parametric estimation under a class of multivariate distributions. Stat Pap 46:321–338
Al-Hussaini EK, Ateya SF (2006) A class of multivariate distributions and new copulas. J Egypt Math Soc 14:45–54
Al-Hussaini EK, Ateya SF (2012) Bayesian prediction under a class of multivariate distributions. Arab J Math 1:283–293
Ashbacher C (2014) Introduction to neutrosophic logic. American Research Press and Charles Ashbacher, Rehoboth
Deepthi KS, Chacko VM (2020) An upside-down bathtub-shaped failure rate model using a DUS transformation of lomax distribution. Stochastic models in reliability engineering. CRC Press, Boca Raton, pp 81–100
Duan W-Q, Khan Z, Gulistan M, Khurshid A (2021) Neutrosophic exponential distribution: modeling and applications for complex data analysis. Complexity 2021:1–8
Elaal MKA, Jarwan RS (2017) Inference of bivariate generalized exponential distribution based on copula functions. Appl Math Sci 11(24):1155–1186
Ghosh S, Rana A, Kansal V (2020) A benchmarking framework using nonlinear manifold detection techniques for software defect prediction. Int J Comput Sci Eng 21(4):593–614
Granados C, Das AK, Birojit D (2022) Some continuous neutrosophic distributions with neutrosophic parameters based on neutrosophic random variables. Adv Theory Nonlinear Anal Appl 6(3):380–389
Hassan MK (2022) Ranked set sampling on estimation of P[Y<X] for inverse Weibull distribution and its applications. Int J Qual Reliab Manag 39(7):1535–1550
Karakaya KKI (2021) On the DUS-Kumaraswamy distribution. Istatistik J Turk Stat Assoc 13:29–38
Khan Z, Gulistan M, Kausar N, Park C (2021) Neutrosophic Rayleigh model with some basic characteristics and engineering applications. IEEE Access 9:71277–71283
Kotz S, Balakrishnan N, Johnson NL (2000) Continuous multivariate distributions: models and applications, vol 1. Wiley, New York
Kumaraswamy P (1980) A generalized probability density function for double-bounded random processes. J Hydrol 46(1–2):79–88
Kundu D, Howlader H (2010) Bayesian inference and prediction of the inverse Weibull distribution for Type-II censored data. Comput Stat Data Anal 54:1547–1558
Nayana BM, Anakha KK, Chacko VM, Aslam M, Albassam M (2022) A new neutrosophic model using DUS-Weibull transformation with application. Complex Intell Syst 8:4079–4088
Nelsen RB (2006) An introduction to copulas. Springer, New York
Patro SK, Smarandache F (2016) The neutrosophic statistical distribution, more problems, more solutions. Neutrosoph Sets Syst 12
Salem S, Khan Z, Ayed H, Brahmia A, Amin A (2021) The neutrosophic lognormal model in lifetime data analysis: properties and applications. J Funct Sp 202:1–91
Shah F, Aslam M, Khan Z, Almazah M, Alduais FS (2022) On neutrosophic extension of the maxwell model: properties and applications. J Funct Sp 2022:1–9
Sherwani RAK, Arshad T, Albassam M, Aslam M, Abbas S (2021) Neutrosophic entropy measures for the Weibull distribution: theory and applications. Complex Intell Syst 7(6):3067–3076
Sherwani RAK, Naeem M, Aslam M, Raza MA, Abid M, Abbas S (2021) Neutrosophic beta distributionwith properties and applications. Neutrosoph Sets Syst 41:209214
Singh SK, Singh U, Kumar D (2013) Bayesian estimation of parameters of inverse Weibull distribution. J Appl Stat 40(7):1597–1607
Smarandache F (2014) Introduction to neutrosophic statistics. Sitech & Education Publishing, Craiova
Tripathi A, Singh U, Singh SK (2019) Inferences for the dusexponential distribution based on upper record values. Ann Data Sci 8:1–17
Wibawati W, Ahsan M, Khusna H, Qori’atunnadyah M, Udiatami WM (2022) Multivariate control chart based on neutrosophic hotelling T2 statistics and its application. JTAM (Jurnal Teori dan Aplikasi Matematika) 6(1):85–92
Zeina MB, Hatip A (2021) Neutrosophic random variables. Neutrosoph Sets Syst 39(1):44–52
Acknowledgements
The authors are deeply thankful to the editor and reviewers for their valuable suggestions to improve the quality and presentation of the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No conflict of interest regarding this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hassan, M.K.H., Aslam, M. DUS-neutrosophic multivariate inverse Weibull distribution: properties and applications. Complex Intell. Syst. 9, 5679–5691 (2023). https://doi.org/10.1007/s40747-023-01026-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-023-01026-2