Abstract
Purpose of Review
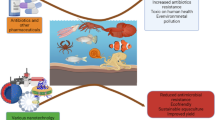
Triclosan (TCS), a widely used broad-spectrum antimicrobial agent, enters to wastewater treatment plants (WWTPs) and the environment ultimately after its usage. Notably, the use of TCS has surged during the outbreak of COVID-19, leading to the environment under increasing TCS pollution pressure. Even environmentally relevant concentrations of TCS can promote the development of antimicrobial resistance (AMR), which is a major public health concern. The purpose of this study is to provide a basis for the management and risk assessment of TCS by providing a holistic review of the impact of TCS on AMR in the environment.
Recent Findings
Bacterial resistance to TCS mainly takes place through modification or replacement of the FabI enzyme, which is the main target of TCS in bacteria. Currently, multiple FabI mutants and isoenzymes have been identified in the environment giving bacterial resistance to TCS. In addition, mechanisms by which TCS promotes bacterial development of resistance to other antimicrobials have been studied in laboratory experiments and environmental settings, such as anaerobic digester. TCS will promote the development of AMR in the environment with the possibility of adverse risks to public health.
Summary
This review systematically summarizes the mechanisms of bacterial resistance to antimicrobials driven by TCS and highlights the effects of TCS in promoting the horizontal transfer and enrichment of antibiotic resistance genes (ARGs). Suggestions for overcoming the limitations of laboratory-scale studies and further improving the risk assessment of TCS in the environment are proposed.



Similar content being viewed by others
References
Sabaliunas D, Webb SF, Hauk A, Jacob M, Eckhoff WS. Environmental fate of triclosan in the river Aire basin. UK Water Res. 2003;37(13):3145–54. https://doi.org/10.1016/S0043-1354(03)00164-7.
Bester K. Triclosan in a sewage treatment process - balances and monitoring data. Water Res. 2003;37(16):3891–6. https://doi.org/10.1016/S0043-1354(03)00335-X.
Zheng X, Yan Z, Liu P, Fan J, Wang S, Wang P, et al. Research progress on toxic effects and water quality criteria of triclosan. Bull Environ Contam Toxicol. 2019;102(6):731–40. https://doi.org/10.1007/s00128-019-02603-3.
Weatherly LM, Gosse JA. Triclosan exposure, transformation, and human health effects. J Toxicol Environ Health B Crit Rev. 2017;20(8):447–69. https://doi.org/10.1080/10937404.2017.1399306.
Zhang J, Walker ME, Sanidad KZ, Zhang H, Liang Y, Zhao E, et al. Microbial enzymes induce colitis by reactivating triclosan in the mouse gastrointestinal tract. Nat Commun. 2022;13(1):136. https://doi.org/10.1038/s41467-021-27762-y.
Stoker TE, Gibson EK, Zorrilla LM. Triclosan exposure modulates estrogen-dependent responses in the female wistar rat. Toxicol Sci. 2010;117(1):45–53. https://doi.org/10.1093/toxsci/kfq180.
FDA (U.S. Food and Drug Administration). 21 CFR part 310 safety and effectiveness of consumer antiseptics. topical antimicrobial drug products for over-the-counter human use. Final rule. Fed Reg. 2016;81:61106–61130.
ECHA (European Chemicals Agency). Biocidal products committee (BPC): opinion on the application for approval of the active substance: triclosan product-type: 1. 2015. Available online: https://echa.europa.eu/documents/10162/efc985e4–8802–4ebb-8245–29708747a358. (Accessed on 17 June 2016).
Guerra P, Teslic S, Shah A, Albert A, Gewurtz SB, Smyth SA. Occurrence and removal of triclosan in Canadian wastewater systems. Environ Sci Pollut Res Int. 2019;26(31):31873–86. https://doi.org/10.1007/s11356-019-06338-w.
Peng FJ, Pan CG, Zhang M, Zhang NS, Windfeld R, Salvito D, et al. Occurrence and ecological risk assessment of emerging organic chemicals in urban rivers: Guangzhou as a case study in China. Sci Total Environ. 2017;589:46–55. https://doi.org/10.1016/j.scitotenv.2017.02.200.
Chen ZF, Wen HB, Dai X, Yan SC, Zhang H, Chen YY, et al. Contamination and risk profiles of triclosan and triclocarban in sediments from a less urbanized region in China. J Hazard Mater. 2018;357:376–83. https://doi.org/10.1016/j.jhazmat.2018.06.020.
•• Barrett H, Sun J, Gong Y, Yang P, Hao C, Verreault J, et al. Triclosan is the predominant antibacterial compound in ontario sewage sludge. Environ Sci Technol. 2022. https://doi.org/10.1021/acs.est.2c00406. This study highlighted TCS as the predominant antibacterial compound in sewage sludge impacting E. coli.
Shi X, Xia Y, Wei W, Ni BJ. Accelerated spread of antibiotic resistance genes (ARGs) induced by non-antibiotic conditions: roles and mechanisms. Water Res. 2022;224:119060. https://doi.org/10.1016/j.watres.2022.119060.
https://www.epa.gov/sites/default/files/202006/documents/sars-cov2_listn_06122020.pdf.
Zhang Y, Walsh TR, Wang Y, Shen JZ, Yang M. Minimizing risks of antimicrobial resistance development in the environment from a public one health perspective. China CDC Wkly. 2022;4(49):1105–9. https://doi.org/10.46234/ccdcw2022.224.
• Carey DE, McNamara PJ. The impact of triclosan on the spread of antibiotic resistance in the environment. Front Microbiol. 2014;5:780. https://doi.org/10.3389/fmicb.2014.00780. This review focused on the effects of TCS on antimicrobial resistance in the environment, including engineered environments such as anaerobic digesters.
Kummerer K. Resistance in the environment. J Antimicrob Chemother. 2004;54(2):311–20. https://doi.org/10.1093/jac/dkh325.
Gerald M. Antiseptics and disinfectants: activity, action, and resistance. Clinical Microbiology Rev. 1999;12:147–79.
Dai S, Liu D, Han Z, Wang Y, Lu X, Yang M, et al. Mobile tigecycline resistance gene tet(X4) persists with different animal manure composting treatments and fertilizer receiving soils. Chemosphere. 2022;307(Pt 3):135866. https://doi.org/10.1016/j.chemosphere.2022.135866.
Lucien MAB, Canarie MF, Kilgore PE, Jean-Denis G, Fenelon N, Pierre M, et al. Antibiotics and antimicrobial resistance in the COVID-19 era: Perspective from resource-limited settings. Int J Infect Dis. 2021;104:250–4. https://doi.org/10.1016/j.ijid.2020.12.087.
Carlet J, Jarlier V, Harbarth S, Voss A, Goossens H, Pittet D. Ready for a world without antibiotics? The Pensières Antibiotic Resistance Call to Action. Antimicrob Resist In. 2012;1(1):11. https://doi.org/10.1186/2047-2994-1-11.
• Lu J, Yu Z, Ding P, Guo J. Triclosan promotes conjugative transfer of antibiotic resistance genes to opportunistic pathogens in environmental microbiome. Environ Sci Technol. 2022. https://doi.org/10.1021/acs.est.2c05537. This article demonstrated that environmentally concentrations of TCS significantly enhanced the conjugative transfer of the RP4 plasmid among activated sludge communities, and even mediated the transfer of the RP4 plasmid to opportunistic human pathogens.
Al-Sayah MH. Chemical disinfectants of COVID-19: an overview. J Water Health. 2020;18(5):843–8. https://doi.org/10.2166/wh.2020.108.
Lu J, Guo J. Disinfection spreads antimicrobial resistance Science. 2021;371(6528):474. https://doi.org/10.1126/science.abg4380.
Giuliano CA, Rybak MJ. Efficacy of triclosan as an antimicrobial hand soap and its potential impact on antimicrobial resistance: a focused review. Pharmacotherapy. 2015;35(3):328–36. https://doi.org/10.1002/phar.1553.
Siamak P, Yazdankhah AAS. Triclosan and antimicrobial resistance in bacteria: an overview. Microb Drug Resist. 2006;12:83–9. https://doi.org/10.1089/mdr.2006.12.83.
Aiello AE, Larson E. Antibacterial cleaning and hygiene products as an emerging risk factor for antibiotic resistance in the community. Lancet Infect Dis. 2003;3(8):501–6. https://doi.org/10.1016/S1473-3099(03)00723-0.
Shrestha P, Zhang Y, Chen WJ, Wong TY. Triclosan: antimicrobial mechanisms, antibiotics interactions, clinical applications, and human health. J Environ Sci Health C Toxicol Carcinog. 2020;38(3):245–68. https://doi.org/10.1080/26896583.2020.1809286.
Wang Y, Liang W. Occurrence, toxicity, and removal methods of triclosan: a timely review. Curr Pollut Rep. 2021;7(1):31–9. https://doi.org/10.1007/s40726-021-00173-9.
Wang M, Hu B, Zhou W, Huang K, Fu J, Zhang A, et al. Enhanced hand-to-mouth exposure from hand sanitizers during the COVID-19 pandemic: a case study of triclosan. Sci Bull. 2022;67(10):995–8. https://doi.org/10.1016/j.scib.2022.03.016.
CEO of Lysol maker: Sales are up as a result of coronavirus pandemic [WWW Document],n.d.CEO of Lysol maker: Sales are up as a result of coronavirus pandemic [WWW Doc-ument], n.d. URL https://www.cnbc.com/2020/07/16/ceo-of-durex-condom-maker-intimate-occasions-down-during-pandemic.html.
Crawford BR, deCatanzaro D. Disruption of blastocyst implantation by triclosan in mice: impacts of repeated and acute doses and combination with bisphenol-A. Reprod Toxicol. 2012;34(4):607–13. https://doi.org/10.1016/j.reprotox.2012.09.008.
Ying GG, Yu XY, Kookana RS. Biological degradation of triclocarban and triclosan in a soil under aerobic and anaerobic conditions and comparison with environmental fate modelling. Environ Pollut. 2007;150(3):300–5. https://doi.org/10.1016/j.envpol.2007.02.013.
Heidler J, Halden RU. Meta-analysis of mass balances examining chemical fate during wastewater treatment. Environ Sci Technol. 2008;42(17):6324–32. https://doi.org/10.1021/es703008y.
Middleton JH, Salierno JD. Antibiotic resistance in triclosan tolerant fecal coliforms isolated from surface waters near wastewater treatment plant outflows (Morris County, NJ, USA). Ecotoxicol Environ Saf. 2013;88:79–88. https://doi.org/10.1016/j.ecoenv.2012.10.025.
Adhikari S, Kumar R, Driver EM, Perleberg TD, Yanez A, Johnston B, et al. Mass trends of parabens, triclocarban and triclosan in Arizona wastewater collected after the 2017 FDA ban on antimicrobials and during the COVID-19 pandemic. Water Res. 2022;222:118894. https://doi.org/10.1016/j.watres.2022.118894.
Ramaswamy BS, Velu G, Rengarajan G, Larsson G, Joakim DG. GC-MS analysis and ecotoxicological risk assessment of triclosan, carbamazepine and parabens in Indian rivers. J Hazard Mater. 2011;186(2–3):1586–93. https://doi.org/10.1016/j.jhazmat.2010.12.037.
Wang XK, Jiang XJ, Wang YN, Sun J, Wang C, Shen TT. Occurrence, distribution, and multi-phase partitioning of triclocarban and triclosan in an urban river receiving wastewater treatment plants effluent in China. Environ Sci Pollut R. 2014;21(11):7065–74. https://doi.org/10.1007/s11356-014-2617-1.
Walters E, McClellan K, Halden RU. Occurrence and loss over three years of 72 pharmaceuticals and personal care products from biosolids-soil mixtures in outdoor mesocosms. Water Res. 2010;44(20):6011–20. https://doi.org/10.1016/j.watres.2010.07.051.
Huang XL, Wu CX, Xiong X, Zhang K, Liu JT. Partitioning and degradation of triclosan and formation of methyl-triclosan in water-sediment systems. Water Air Soil Pollut. 2014;225(9). https://doi.org/10.1007/s11270-014-2099-2.
• Guo J, Iwata H. Risk assessment of triclosan in the global environment using a probabilistic approach. Ecotoxicol Environ Saf. 2017;143:111–9. https://doi.org/10.1016/j.ecoenv.2017.05.020. This article collected data on the concentration of TCS in the environment and makes a risk assessment of TCS.
Juksu K, Zhao JL, Liu YS, Yao L, Sarin C, Sreesai S, et al. Occurrence, fate and risk assessment of biocides in wastewater treatment plants and aquatic environments in Thailand. Sci Total Environ. 2019;690:1110–9. https://doi.org/10.1016/j.scitotenv.2019.07.097.
Lalonde B, Garron C, Dove A, Struger J, Farmer K, Sekela M, et al. Investigation of spatial distributions and temporal trends of triclosan in Canadian surface waters. Arch Environ Contam Toxicol. 2019;76(2):231–45. https://doi.org/10.1007/s00244-018-0576-0.
Zhu Q, Jia J, Wang Y, Zhang K, Zhang H, Liao C, et al. Spatial distribution of parabens, triclocarban, triclosan, bisphenols, and tetrabromobisphenol A and its alternatives in municipal sewage sludges in China. Sci Total Environ. 2019;679:61–9. https://doi.org/10.1016/j.scitotenv.2019.05.059.
McMurry L, Oethinger M, Levy S. Triclosan targets lipid synthesis Nat. 1998;394:531–2.
Lu YJ, Zhang YM, Rock CO. Product diversity and regulation of type II fatty acid synthases. Biochem Cell Biol. 2004;82(1):145–55. https://doi.org/10.1139/o03-076.
Zhu L, Lin J, Ma J, Cronan JE, Wang H. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoyl-acyl carrier protein reductase. Antimicrob Agents Chemother. 2010;54(2):689–98. https://doi.org/10.1128/AAC.01152-09.
Yu Y, Ma J, Wang H. Advances in fatty acid biosynthetic diversity in bacteria. J Microbiol. 2016;36(4):76–83.
Pidugu LS, Kapoor M, Surolia N, Surolia A, Suguna K. Structural basis for the variation in triclosan affinity to enoyl reductases. J Mol Biol. 2004;343(1):147–55. https://doi.org/10.1016/j.jmb.2004.08.033.
Levy C, Roujeinikova A, Sedelnikova S. Molecular basis of triclosan activity Nat. 1999;398:383. https://doi.org/10.1038/18803.
•• Drury B, Scott J, Rosi-Marshall EJ, Kelly JJ. Triclosan exposure increases triclosan resistance and influences taxonomic composition of benthic bacterial communities. Environ Sci Technol. 2013;47(15):8923–30. https://doi.org/10.1021/es401919k.This study demonstrated the effect of TCS on the composition and resistance of benthic bacterial communities in field and artificial streams.
Morvan C, Halpern D, Kenanian G, Pathania A, Anba-Mondoloni J, Lamberet G, et al. The Staphylococcus aureus FASII bypass escape route from FASII inhibitors. Biochimie. 2017;141:40–6. https://doi.org/10.1016/j.biochi.2017.07.004.
Kim SH, Khan R, Choi K, Lee SW, Rhee S. A triclosan-resistance protein from the soil metagenome is a novel enoyl-acyl carrier protein reductase: structure-guided functional analysis. FEBS J. 2020;287(21):4710–28. https://doi.org/10.1111/febs.15267.
Morvan C, Halpern D, Kenanian G, Hays C, Anba-Mondoloni J, Brinster S, et al. Environmental fatty acids enable emergence of infectious Staphylococcus aureus resistant to FASII-targeted antimicrobials. Nat Commun. 2016;7:12944. https://doi.org/10.1038/ncomms12944.
Fan F, Yan K, Wallis NG, Reed S, Moore TD, Rittenhouse SF, et al. Defining and combating the mechanisms of triclosan resistance in clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(11):3343–7. https://doi.org/10.1128/AAC.46.11.3343-3347.2002.
Supathep T, Liam JR, Gianmarco C, Li Chin W, Kimie R, Adam PR. Reduced susceptibility to antiseptics is conferred by heterologous housekeeping genes. Microb Drug Resist. 2018;24(2):105–12. https://doi.org/10.1089/mdr.2017.0105.
Yu K, Zhang Y, Xu W, Zhang X, Xu Y, Sun Y, et al. Hyper-expression of the efflux pump gene adeB was found in Acinetobacter baumannii with decreased triclosan susceptibility. J Glob Antimicrob Resist. 2020;22:367–73. https://doi.org/10.1016/j.jgar.2020.02.027.
• Ciusa ML, Furi L, Knight D, Decorosi F, Fondi M, Raggi C, et al. A novel resistance mechanism to triclosan that suggests horizontal gene transfer and demonstrates a potential selective pressure for reduced biocide susceptibility in clinical strains of Staphylococcus aureus. Int J Antimicrob Agents. 2012;40(3):210–20. https://doi.org/10.1016/j.ijantimicag.2012.04.021. This paper described sh-fabI as a novel resistance mechanism with high potential for horizontal gene transfer.
Yu BJ, Kim JA, Pan J-G. Signature gene expression profile of triclosan-resistant Escherichia coli. J Antimicrob Chemother. 2010;65(6):1171–7. https://doi.org/10.1093/jac/dkq114.
Chen Y, Pi B, Zhou H, Yu Y, Li L. Triclosan resistance in clinical isolates of Acinetobacter baumannii. J Med Microbiol. 2009;58(Pt 8):1086–91. https://doi.org/10.1099/jmm.0.008524-0.
Qiu XY, Janson CA, Court RI, Smyth MG, Payne DJ, Abdel-Meguid SS. Molecular basis for triclosan activity involves a flipping loop in the active site. Protein Sci. 1999;8(11):2529–32. https://doi.org/10.1110/ps.8.11.2529.
Stewart MJ, Parikh S, Xiao GP, Tonge PJ, Kisker C. Structural basis and mechanism of enoyl reductase inhibition by triclosan. J Mol Biol. 1999;290(4):859–65. https://doi.org/10.1006/jmbi.1999.2907.
Sivaraman S, Zwahlen J, Bell AF, Hedstrom L, Tonge PJ. Structure-activity studies of the inhibition of FabI, the enoyl reductase from Escherichia coli, by triclosan: kinetic analysis of mutant. Biochemistry. 2003;42(15):4406–13. https://doi.org/10.1021/bi0300229.
•• Khan R, Kong HG, Jung YH, Choi J, Baek KY, Hwang EC, et al. Triclosan resistome from metagenome reveals diverse enoyl acyl carrier protein reductases and selective enrichment of triclosan resistance genes. Sci Rep. 2016;6:32322. https://doi.org/10.1038/srep32322. This paper showed the TCS-resistant ENRs and TCS resistance genes in soil using metagenome.
Demissie R, Kabre P, Fung LW. Nonactive-site mutations in S. aureus FabI that induce triclosan resistance. ACS Omega. 2020;5(36):23175–83. https://doi.org/10.1021/acsomega.0c02942.
Heath RJ, Su N, Murphy CK, Rock CO. The enoyl-[acyl-carrier-protein] reductases FabI and FabL from Bacillus subtilis. J Biol Chem. 2000;275(51):40128–33. https://doi.org/10.1074/jbc.M005611200.
Massengo-Tiasse RP, Cronan JE. Vibrio cholerae FabV defines a new class of enoyl-acyl carrier protein reductase. J Biol Chem. 2008;283(3):1308–16. https://doi.org/10.1074/jbc.M708171200.
•• Khan AU, Cameron A, Barbieri R, Read R, Church D, Adator EH, et al. Functional screening for triclosan resistance in a wastewater metagenome and isolates of Escherichia coli and Enterococcus spp. from a large Canadian healthcare region. Plos One. 2019;14(1). https://doi.org/10.1371/journal.pone.0211144. This article identified functional mechanisms of TCS resistance in waste water metagenomes, and assessed the frequency of TCS resistance in waste water-derived and clinical isolates.
Alexander G, McFarland HKB. Triclosan tolerance is driven by a conserved mechanism in diverse pseudomonas species. Appl Environ Microbiol. 2021;87(7):e02924-e3020. https://doi.org/10.1128/AEM.02924-20.
Hopf FSM, Roth CD, de Souza EV, Galina L, Czeczot AM, Machado P, et al. Bacterial enoyl-reductases: the ever-growing list of fabs, their mechanisms and inhibition. Front Microbiol. 2022;13:891610. https://doi.org/10.3389/fmicb.2022.891610.
Heath R, Rock C. A triclosan-resistant bacterial enzyme Nat. 2000;406:145. https://doi.org/10.1038/35022656.
Khan R, Zeb A, Roy N, Thapa Magar R, Kim HJ, Lee KW, et al. Biochemical and structural basis of triclosan resistance in a novel enoyl-acyl carrier protein reductase. Antimicrob Agents Chemother. 2018;62(8). https://doi.org/10.1128/AAC.00648-18.
Khan R, Zeb A, Choi K, Lee G, Lee KW, Lee SW. Biochemical and structural insights concerning triclosan resistance in a novel YX7K type enoyl-acyl carrier protein reductase from soil metagenome. Sci Rep. 2019;9(1):15401. https://doi.org/10.1038/s41598-019-51895-2.
Huang YH, Lin JS, Ma JC, Wang HH. Functional characterization of triclosan-resistant enoyl-acyl-carrier protein reductase (FabV) in Pseudomonas aeruginosa. Front Microbiol. 2016;7:1903. https://doi.org/10.3389/fmicb.2016.01903.
Zhu L, Bi H, Ma J, Hu Z. The two functional enoyl-acyl carrier protein reductases of Enterococcus faecalis do not mediate triclosan resistance. MBIO. 2013;4(5). https://doi.org/10.1128/mBio.00613-13.
Taubes G. The bacteria fight back. Sci Total Environ. 2008;321(5887):356–61. https://doi.org/10.1126/science.321.5887.356.
• Cave R, Cole J, Mkrtchyan HV. Surveillance and prevalence of antimicrobial resistant bacteria from public settings within urban built environments: challenges and opportunities for hygiene and infection control. Environ Int. 2021;157:106836. https://doi.org/10.1016/j.envint.2021.106836. This study showed that a low abundance of antibiotic resistance in public settings compared with environments where stronger disinfectants are used though metagenomic analysis.
Yu Zhang JX, Miaomiao Liu. Microbial community functional structure in response to antibiotics in pharmaceutical wastewater treatment systems. Water Res. 2013;47:6298–308. https://doi.org/10.1016/j.watres.2013.08.003.
Yu X, Zuo JN, Li RX, Gan LL, Li ZX, Zhang F. A combined evaluation of the characteristics and acute toxicity of antibiotic wastewater. Ecotoxicol Environ Saf. 2014;106:40–5. https://doi.org/10.1016/j.ecoenv.2014.04.035.
Li D, Gao J, Dai H, Wang Z, Duan W. Long-term responses of antibiotic resistance genes under high concentration of enrofloxacin, sulfadiazine and triclosan in aerobic granular sludge system. Bioresour Technol. 2020;312:123567. https://doi.org/10.1016/j.biortech.2020.123567.
Zhao JL, Ying GG, Liu YS, Chen F, Yang JF, Wang L. Occurrence and risks of triclosan and triclocarban in the Pearl River system, South China: from source to the receiving environment. J Hazard Mater. 2010;179(1–3):215–22. https://doi.org/10.1016/j.jhazmat.2010.02.082.
Alexandra C, Daniella A, M. Elias D, Michiel V, Gabriel G P. Triclosan alters microbial communities in freshwater microcosms. Water. 2019;11(5). https://doi.org/10.3390/w11050961.
•• Dai H, Gao J, Li D, Wang Z, Zhao Y, Cui Y. Polyvinyl chloride microplastics changed risks of antibiotic resistance genes propagation by enhancing the removal of triclosan in partial denitrification systems with different carbon source. Chem Eng J. 2022;429. https://doi.org/10.1016/j.cej.2021.132465. This research article demonstrated that TCS affected the distribution of ARGs in the environment by altering the microbial community structure.
Carey DE, McNamara PJ. Altered antibiotic tolerance in anaerobic digesters acclimated to triclosan or triclocarban. Chemosphere. 2016;163:22–6. https://doi.org/10.1016/j.chemosphere.2016.07.097.
Li D, Gao J, Dai H, Wang Z, Cui Y, Zhao Y, et al. Triclosan enriched resistance genes more easily than copper in the presence of environmental tetracycline in aerobic granular sludge system. Sci Total Environ. 2022;815:152871. https://doi.org/10.1016/j.scitotenv.2021.152871.
Wolters B, Hauschild K, Blau K, Mulder I, Heyde BJ, Sorensen SJ, et al. Biosolids for safe land application: does wastewater treatment plant size matters when considering antibiotics, pollutants, microbiome, mobile genetic elements and associated resistance genes? Environ Microbiol. 2022;24(3):1573–89. https://doi.org/10.1111/1462-2920.15938.
Fujimoto M, Carey DE, McNamara PJ. Metagenomics reveal triclosan-induced changes in the antibiotic resistome of anaerobic digesters. Environ Pollut. 2018;241:1182–90. https://doi.org/10.1016/j.envpol.2018.06.048.
Gao JF, Liu XH, Fan XY, Dai HH. Effects of triclosan on performance, microbial community and antibiotic resistance genes during partial denitrification in a sequencing moving bed biofilm reactor. Bioresour Technol. 2019;281:326–34. https://doi.org/10.1016/j.biortech.2019.02.112.
Oh S, Choi D, Cha CJ. Ecological processes underpinning microbial community structure during exposure to subinhibitory level of triclosan. Sci Rep. 2019;9(1):4598. https://doi.org/10.1038/s41598-019-40936-5.
Zhao Y, Gao J, Zhang W, Wang Z, Cui Y, Dai H, et al. Robustness of the partial nitrification-anammox system exposing to triclosan wastewater: stress relieved by extracellular polymeric substances and resistance genes. Environ Res. 2022;206:112606. https://doi.org/10.1016/j.envres.2021.112606.
Tan Q, Chen J, Chu Y, Liu W, Yang L, Ma L, et al. Triclosan weakens the nitrification process of activated sludge and increases the risk of the spread of antibiotic resistance genes. J Hazard Mater. 2021;416:126085. https://doi.org/10.1016/j.jhazmat.2021.126085.
Han Y, Zhou ZC, Zhu L, Wei YY, Feng WQ, Xu L, et al. The impact and mechanism of quaternary ammonium compounds on the transmission of antibiotic resistance genes. Environ Sci Pollut Res Int. 2019;26(27):28352–60. https://doi.org/10.1007/s11356-019-05673-2.
Thomas CM, Nielsen KM. Mechanisms of, and Barriers to, Horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3(9):711–21. https://doi.org/10.1038/nrmicro1234.
Jutkina J, Marathe NP, Flach CF, Larsson DGJ. Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci Total Environ. 2018;616–617:172–8. https://doi.org/10.1016/j.scitotenv.2017.10.312.
Lu J, Wang Y, Li J, Mao L, Nguyen SH, Duarte T, et al. Triclosan at environmentally relevant concentrations promotes horizontal transfer of multidrug resistance genes within and across bacterial genera. Environ Int. 2018;121(Pt 2):1217–26. https://doi.org/10.1016/j.envint.2018.10.040.
Lu J, Wang Y, Zhang S, Bond P, Yuan Z, Guo J. Triclosan at environmental concentrations can enhance the spread of extracellular antibiotic resistance genes through transformation. Sci Total Environ. 2020;713:136621. https://doi.org/10.1016/j.scitotenv.2020.136621.
Lu Y, Zeng JM, Wang LJ, Lan K, E SM, Wang LN, et al. Antibiotics promote escherichia coli-pseudomonas aeruginosa conjugation through inhibiting quorum sensing. Antimicrob Agents Chemother. 2017;61(12). https://doi.org/10.1128/AAC.01284-17.
Puangseree J, Jeamsripong S, Prathan R, Pungpian C, Chuanchuen R. Resistance to widely-used disinfectants and heavy metals and cross resistance to antibiotics in Escherichia coli isolated from pigs, pork and pig carcass. Food Control. 2021;124. https://doi.org/10.1016/j.foodcont.2021.107892.
Lin F, Xu Y, Chang Y, Liu C, Jia X, Ling B. Molecular characterization of reduced susceptibility to biocides in clinical isolates of Acinetobacter baumannii. Front Microbiol. 2017;8:1836. https://doi.org/10.3389/fmicb.2017.01836.
Hernandez A, Ruiz FM, Romero A, Martinez JL. The binding of triclosan to SmeT, the repressor of the multidrug efflux pump SmeDEF, induces antibiotic resistance in Stenotrophomonas maltophilia. PLoS Pathog. 2011;7(6):e1002103. https://doi.org/10.1371/journal.ppat.1002103.
Chuanchuen R, Beinlich K, Hoang TT, Becher A, Karkhoff-Schweizer RR, Schweizer HP. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob Agents Chemother. 2001;45(2):428–32. https://doi.org/10.1128/AAC.45.2.428-432.2001.
Nontaleerak B, Tasnawijitwong N, Eurtivong C, Sirikanchana K, Satayavivad J, Sukchawalit R, et al. Characterisation of the triclosan efflux pump TriABC and its regulator TriR in Agrobacterium tumefaciens C58. Microbiol Res. 2022;263:127112. https://doi.org/10.1016/j.micres.2022.127112.
Lu J, Jin M, Nguyen SH, Mao L, Li J, Coin LJM, et al. Non-antibiotic antimicrobial triclosan induces multiple antibiotic resistance through genetic mutation. Environ Int. 2018;118:257–65. https://doi.org/10.1016/j.envint.2018.06.004.
Lv L, Wan M, Wang C, Gao X, Yang Q, Partridge SR, et al. Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in Klebsiella pneumoniae. MBIO. 2020;11(2). https://doi.org/10.1128/mBio.02930-19.
Pumbwe L, Ueda O, Yoshimura F, Chang A, Smith RL, Wexler HM. Bacteroides fragilis BmeABC efflux systems additively confer intrinsic antimicrobial resistance. J Antimicrob Chemother. 2006;58(1):37–46. https://doi.org/10.1093/jac/dkl202.
Lin J, Michel LO, Zhang QJ. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother. 2002;46(7):2124–31. https://doi.org/10.1128/AAC.46.7.2124-2131.2002.
Hansen LH, Jensen LB, Sørensen HI, Sørensen SJ. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother. 2007;60(1):145–7. https://doi.org/10.1093/jac/dkm167.
Rodriguez-Martinez JM, Diaz de Alba P, Briales A, Machuca J, Lossa M, Fernandez-Cuenca F, et al. Contribution of OqxAB efflux pumps to quinolone resistance in extended-spectrum-lactamase-producing Klebsiella pneumoniae. J Antimicrob Chemoth. 2012;68(1):68–73. https://doi.org/10.1093/jac/dks377.
Milani ES, Hasani A, Varschochi M, Sadeghi J, Memar MY, Hasani A. Biocide resistance in Acinetobacter baumannii: appraising the mechanisms. J Hosp Infect. 2021. https://doi.org/10.1016/j.jhin.2021.10.009.
Henly EL, Dowling JAR, Maingay JB, Lacey MM, Smith TJ, Forbes S. Biocide exposure induces changes in susceptibility, pathogenicity, and biofilm formation in uropathogenic Escherichia coli. Antimicrob Agents Chemother. 2019;63(3). https://doi.org/10.1128/AAC.01892-18.
• Li M, He Y, Sun J, Li J, Bai J, Zhang C. Chronic exposure to an environmentally relevant triclosan concentration induces persistent triclosan resistance but reversible antibiotic tolerance in Escherichia coli. Environ Sci Technol. 2019;53(6):3277–86. https://doi.org/10.1021/acs.est.8b06763. This research article suggested that biofilm augmentation was the main reason behind the development of bacterial resistance to antimicrobials.
Westfall C, Flores-Mireles A, Robinson J, Lynch AJL, Hultgren S, Henderson J, et al. The widely used antimicrobial triclosan induces high levels of antibiotic tolerance in vitro and reduces antibiotic efficacy up to 100-fold in vivo. Antimicrob Agents Chemother. 2019;63(5). https://doi.org/10.1128/aac.02312-18.
Harms A, Fino C, Sorensen MA, Semsey S, Gerdes K. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. MBIO. 2017;8(6). https://doi.org/10.1128/mBio.01964-17.
Seyfzadeh M, Keener J, Nomura M. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc Natl Acad Sci. 1993;90(23):11004–8. https://doi.org/10.1073/pnas.90.23.11004.
Korch SB, Henderson TA, Hill TM. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol Microbiol. 2003;50(4):1199–213. https://doi.org/10.1046/j.1365-2958.2003.03779.x.
Viducic D, Ono T, Murakami K, Susilowati H, Kayama S, Hirota K, et al. Functional analysis of spoT, relA and dksA genes on quinolone tolerance in Pseudomonas aeruginosa under nongrowing condition. Med Microbiol Immunol. 2006;50(4):349–57. https://doi.org/10.1111/j.1348-0421.2006.tb03793.x.
Battesti A, Bouveret E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol. 2006;62(4):1048–63. https://doi.org/10.1111/j.1365-2958.2006.05442.x.
Hoang TT, Sullivan SA, Cusick JK, Schweizer HP. β-ketoacyl acyl carrier protein reductase (FabG) activity of the fatty acid biosynthetic pathway is a determining factor of 3-oxo-homoserine lactone acyl chain lengths. MicrobiologyI-SGM. 2002;148:3849–56. https://doi.org/10.1099/00221287-148-12-3849.
Zhao X, Yu Z, Ding T. Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms. 2020;8(3). https://doi.org/10.3390/microorganisms8030425.
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Tong C, Hu H, Chen G, Li Z, Li A, Zhang J. Disinfectant resistance in bacteria: mechanisms, spread, and resolution strategies. Environ Res. 2021;195:110897. https://doi.org/10.1016/j.envres.2021.110897.
Zhang K, Hong H. Mechanisms of microbial disinfectant resistance. Prog Biochem Biophys. 2022;49:34–47. https://doi.org/10.16476/j.pibb.2021.0300. (in Chinese).
Funding
This work was supported by the National Natural Science Foundation of China (Grants 32141002).
Author information
Authors and Affiliations
Contributions
Chunzhen Wang wrote the main manuscript text; Shihai Liu and Haodi Feng prepared figures 1-2 and edited the manuscript; Yu Zhang drafted original manuscript and provided funding and supervision; All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, C., Liu, S., Feng, H. et al. Effects of Triclosan on the Development of Antimicrobial Resistance in the Environment: A Review. Curr Pollution Rep 9, 454–467 (2023). https://doi.org/10.1007/s40726-023-00270-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40726-023-00270-x