Abstract
Introduction
Imatinib mesylate (IM), a selective inhibitor of the BCR-ABL tyrosine kinase, is a well-established first-line treatment for chronic myeloid leukemia (CML). IM is metabolized mainly by cytochrome P450 (CYP) in the liver, specifically the CYP3A4 and CYP3A5 enzymes. Polymorphisms in these genes can alter the enzyme activity of IM and may affect its response. In this study, the impact of two single-nucleotide polymorphisms (SNPs), CYP3A5*3 (6986A>G) and CYP3A4*18 (878T>C), on IM treatment response in CML patients (n = 270; 139 IM resistant and 131 IM good responders) was investigated.
Methods
Genotyping of CYP3A4*18 and CYP3A5*3 was performed using the polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) technique. The association between allelic variants and treatment response was assessed by means of odds ratio (OR) with 95% confidence intervals calculated by logistic regression.
Results
Our results indicated that CML patients carrying the heterozygous (AG) and homozygous variant (GG) genotype of CYP3A5*3 were associated with a significantly lower risk of acquiring resistance with OR 0.171; 95% CI: 0.090–0.324, p < 0.001 and OR 0.257; 95% CI: 0.126–0.525, p < 0.001, respectively. Although CML patients carrying the heterozygous (TC) genotype of CYP3A4*18 showed a lower risk of acquiring resistance toward IM (OR 0.648; 95% CI: 0.277–1.515), the association was not statistically significant (p = 0.316). No homozygous variant (CC) genotype of CYP3A4*18 was detected among the CML patients.
Conclusion
It is concluded that polymorphism of CYP3A5*3 is associated with IM treatment response in Malaysian CML patients with carriers of CYP3A5*1/*3 and CYP3A5*3/*3 genotypes posing lower risk for development of resistance to IM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Currently, imatinib mesylate (IM) is the frontline therapy for newly diagnosed chronic myeloid leukemia (CML) patients worldwide. IM is a synthetic tyrosine kinase inhibitor specifically designed to inhibit the breakpoint cluster region (BCR)-Abelson (ABL) fusion protein resulting from Philadelphia chromosome translocation t(9; 22)(q34, q11) [1]. Although IM is the gold standard drug for CML treatment, development of resistance to IM is a major clinical problem. The development of resistance to IM is a multifactorial phenomenon in patients with CML. Resistance may be mediated by a range of different mechanisms in which pharmacokinetic variability due to genetic polymorphisms in IM transport and metabolizing genes may be potential factors. Previously, the contribution of genetic variations in transporter genes ABCB1 and ABCG2 in mediating resistance/good response to IM among Malaysian CML patients was reported by our group [2]. Drug-metabolizing enzymes (DME) are involved in deactivating xenobiotics as well as biotransformation of drugs, and polymorphisms in DME coding genes have been reported to alter the activity of the enzymes for some substrates [3]. Therefore, genetic variation in drug metabolism can lead to therapeutic failures, adverse drug effects or even fatal drug intoxications [4].
Xenobiotic metabolism is mainly conducted by three main cytochrome P450 (CYP) gene families: CYP1, CYP2 and CYP3, with the most highly expressed subfamily being the CYP3A. Genetic variation in CYP3A activity may influence the rate of metabolism and elimination of CYP3A substrates including IM. The metabolism of IM is mainly mediated by CYP3A4 and CYP3A5 [5]. It is suggested that CYP3A4 and CYP3A5 are important genetic contributors to inter-individual differences in CYP3A-dependent drug metabolism. A polymorphism of CYP3A4*18, located at exon 10, had been reported as the common allelic variation of CYP3A4. This polymorphism involves nucleotide change from tyrosine (T) to cytosine (C) transition at position 878 and results in an amino acid change from leucine to proline at codon 293 (Leu293Pro) [6, 7]. CYP3A4*18 had been reported to be associated with high turnover of testosterone and chlorpyrifos when compared to the wild type [8]. CYP3A5*3 is another common allelic variation in CYP3A5. Polymorphism in CYP3A5*3 plays a crucial role in the pharmacokinetics of these CYP3A substrates. It is reasonable to assume that polymorphisms in the CYP3A5 gene could lead to interindividual variability, which has been observed in the pharmacokinetic changes seen with CYP3A substrates [9]. This study was designed to investigate the frequency of single-nucleotide polymorphisms (SNPs) CYP3A5*3 (6986A>G) and CYP3A4*18 (878T>C) in Malaysian CML patients undergoing IM therapy and to determine their impact on IM’s treatment response in these patients. To our knowledge, this is the first study to report on the allelic and genotypic frequencies as well as the association of CYP3A4*18 and CYP3A5*3 with IM response among CML patients in the Malaysian population.
Methods
Subjects and DNA Extraction
The study was approved by the Research and Ethics Committee of Universiti Sains Malaysia and the Ministry of Health, Malaysia (ethics no. USMKK/PPP/JEPeM [244.3.(4)] and KKM/NHSEC/08/0804/P12-687), which complies with the Declaration of Helsinki of 1964, as revised in 2013. Informed consent was obtained from all patients for being included in the study. Subjects were recruited from various hospitals in Malaysia including Hospital Universiti Sains Malaysia (HUSM), Hospital Raja Perempuan Zainab II (HRPZ), Hospital Pulau Pinang, Hospital Raja Permaisuri Bainun, Universiti Kebangsaan Malaysia Medical Center (PPUKM), Sime Darby Medical Centre and Hospital Umum Sarawak (HUS). In this study, 270 CML patients (139 IM resistant and 131 IM good responders) were involved who were all BCR-ABL non-mutated. The patients selected were Philadelphia chromosome-positive CML patients in chronic, accelerated or blast phase, treated for at least 12 months, with IM (400 and 600 mg, respectively) on frontline treatment. The patients' characteristics are shown in Table 1.
Evaluation of imatinib response: By referring to European LeukemiaNet recommendations for the management of chronic myeloid leukemia 2013 [10], hematologic, cytogenetic and molecular criteria were accessed. Based on this, patients were grouped into IM good responders and IM-resistant CML patients. Hematologic response was considered as complete when the platelet count was <450 × 109/l; white blood cell count <10 × 109/L; differential without immature granulocytes and with <5% basophils and nonpalpable spleen. The cytogenetic response was defined as complete (0% Ph+ metaphases), partial (1–35% Ph+ metaphases), minor (36–65% Ph+ metaphases), minimal (66–95% Ph+ metaphases) and none (>95–100% Ph+ metaphases) [11]. Molecular response was best assessed according to the International Scale (IS) as the ratio of BCR-ABL1% on a log scale, where 10%, 1%, 0.1%, 0.01%, 0.0032% and 0.001% correspond to a decrease of 1, 2, 3, 4, 4.5 and 5 logs, respectively, below the standard baseline that was used in the IRIS study. A BCR-ABL1 expression of ≤0.1% corresponds to major molecular response (MMR) [10]. CML patients who achieved the above-mentioned response criteria were categorized as IM good responders and those who did not achieve the above response criteria within the specified time frame were categorized under the IM non-responders/resistant group.
Peripheral blood (3 ml) was collected after obtaining written informed consents from the subjects. Genomic DNA was extracted using a DNA extraction kit, QIAGEN QIAamp® DNA Blood Mini kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions. Genotyping was conducted at Human Genome Centre, Universiti Sains Malaysia.
Genotyping of CYP3A4*18 Polymorphisms
Genotyping of CYP3A4*18 was performed by using the polymerase chain reaction restriction fragment length polymorphism (PCR–RFLP) technique. Amplification of CYP3A4*18 was performed by using forward (CACATCAGAATGAAACCACC) and reverse (AGAGCCTTCCTACATAGAGTCA) primers. PCR reactions were conducted in a 25-μl volume 1× PCR buffer, 2.0 µM magnesium chloride (MgCl2), 0.5 µM dNTPs, 0.4 µM of each primer and 1.0 U AmpliTaq Gold Polymerase. Denaturation was at 95 °C for 2 min, followed by 35 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s and a final extension step at 72 °C for 5 min. The 450-bp PCR products were electrophoresed on a 2% agarose gel at 100 V for 30 min.
Following PCR amplification, 4 µl of 450-bp PCR products were digested with 1.0 unit of a restriction enzyme (Msp1) for 1 h at 37 °C. The digested PCR products were analyzed by electrophoresis on a 2% agarose gel at 90 V for 50 min. The homozygous wild-type allele (TT) was identified by the presence of an undigested band (450 bp), while the heterozygous allele (TC) was confirmed by the presence of three fragments at 450, 282 and 168 bp. The homozygous variant allele (CC) was identified by the presence of two fragments at 282 and 168 bp.
Genotyping of CYP3A5*3 Polymorphisms
Genotyping of CYP3A5*3 was performed by using a PCR–RFLP. The 293-bp DNA fragment that contains the CYP3A5*3 allele was amplified with the primer pair 5′-GGTCCAAACAGGGAAGAAATA-3′ (forward) and 5′-CATGACTTAGTAGACAGATGAC-3′ (reverse). The PCR reactions were conducted in a 25-µl volume of 1× PCR buffer, 2.0 µM MgCl2, 0.5 µM dNTPs, 0.4 µM of each primer and 1.0 U AmpliTaq Gold Polymerase with a denaturation step of 95 °C for 2 min, followed by 35 cycles at 95 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s and a final extension step at 72 °C for 5 min. The 293-bp PCR products were run on a 2% agarose gel electrophoresis at 100 V for 30 min.
Following PCR amplification, 4 µl of PCR products was digested by restriction enzyme Ssp1 for 15 min at 37 °C. The digested PCR products were analyzed by electrophoresis on a 3% agarose gel. The homozygous wild-type allele (AA) was identified by the presence of 148, 125 and 20 bp, whereas the homozygous variant allele (GG) was confirmed by the presence of fragments of 168- and 125-bp size. The heterozygous variant allele (AG) was identified by the presence of 168-, 148-, 125- and 20-bp fragments.
Direct Sequencing
Following genotyping, a few samples from each different genotype were randomly selected for sequencing to confirm the expected sequences of each genotype. The PCR product was purified by using a QIAquick PCR purification kit (QIAGEN) before sending it to First BASE Laboratories (Kuala Lumpur, Malaysia) for sequencing.
Statistical Analysis
The frequencies of polymorphic genotypes among IM-resistant and good-response CML patients were compared by using the chi-square test (χ 2). The odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using a binary logistic regression to investigate the risk association of genotypes with IM response. All statistical tests were two sided, and statistical significance was determined as p < 0.05. SPSS v.20.0 (SPSS Inc., Chicago, IL, USA) was utilized. Pair-wise linkage disequilibrium (LD) indices (r 2) were determined using Haploview v.4.2 [12].
Results
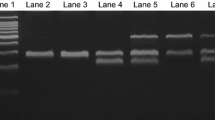
Two hundred seventy (270) CML patients (139 IM resistant and 131 good responders) were successfully recruited. Table 1 shows the patients' characteristics. In the present study, the gender, age and CML stages did not show any influence on IM response (data not shown). In the PCR-RFLP analyses, the restriction enzymes were successfully cut at the correct regions for CYP3A4*18 and CYP3A5*3 (Fig. 1), which was confirmed by the sequencing result (Fig. 2). Our study observed the allelic frequency of CYP3A4*18 and CYP3A5*3 in Malaysian CML patients as 4.44% and 47.41%, respectively. Among the CML patients, the allelic frequency of CYP3A4*18 was lower (3.60%) among the IM-resistant group compared to good responders (5.34%), although this difference was not significant. On the other hand, the allelic frequency of CYP3A5*3 polymorphisms was significantly (p < 0.001) lower (38.85%) among the IM-resistant group compared to the good responders (56.49%).
Gel electrophoresis after RFLP analysis for a CYP3A4*18 (following digestion with Msp1) and b CYP3A5*3 (following digestion with Ssp1). a Lane 1 contained a 100-bp ladder. Lanes 2 and 4 indicated a homozygous wild-type individual. Lane 3 showed a heterozygous individual. b Lane 1 contained a 100-bp ladder. Lanes 2, 3 and 6 showed homozygous variants (GG). Lane 4 indicated a homozygous wild type (AA). Lanes 5 and 7 showed a heterozygous individual. Lane 8 contained a 50-bp ladder
The frequencies of the CYP3A4*18 and CYP3A5*3 genotypes in IM resistant and responders are shown in Table 2. For the CYP3A4*18 polymorphism, only homozygous wild-type (TT) and heterozygous (TC) genotypes were observed among both IM-resistant and good-response CML patients. However, the frequency of CYP3A4*18 homozygous wild type (TT) tended to be higher (92.81%) among the IM-resistant group although this difference was not statistically significant. No homozygous variant (CC) genotype was detected among both IM-resistant and good responders. As for the CYP3A5*3 polymorphism, the genotypic frequencies of the homozygous wild type (AA) were significantly (p < 0.001) higher among the IM-resistant group (44.60% in IM resistant vs. 13.74% in IM good responders), but the frequency of heterozygous genotype (AG) was significantly (p < 0.001) higher in the IM good-responder group (59.54% in IM good responders vs. 33.09% in the IM-resistant group).
To establish the risk association of CYP3A4*18 and CYP3A5*3 polymorphisms with IM response, a binary logistic regression was performed (Table 3 ). For CYP3A4*18, there was no significant association between the polymorphism and the risk of acquiring resistance toward IM. However, for CYP3A5*3 polymorphisms, both heterozygous (AG) and homozygous variants (GG) posed significantly (p < 0.001) lower risk for acquiring resistance toward IM. We determined that these two SNPs, rs28371759 (CYP3A4*18) and rs776746 (CYP3A5*3), are in low linkage disequilibrium (r 2 = 0) probably as a result of recombination occuring between the two markers (Fig. 3).
Discussion
To the best of our knowledge, this is the first study reporting the impact of polymorphism of CYP3A4*18 and CYP3A5*3 in IM treatment response in Malaysian CML patients. For CYP3A4*18, no homozygous variant (*18/*18) was detected among both IM resistant and good responders. This is similar to the findings among the Malaysian population reported by Ruzilawati et al. [13] who did not detect any homozygous variant, indicating that this genotype, which contributes to slower metabolism of most xenobiotics, is rather rare in the Asian population.
CYP3A4*18 was detected at a frequency of 1.7% in a healthy Korean population [14], 2% in a healthy Chinese population [8], 1.3% in a healthy Japanese population [15] and 2.07% among Malaysian diabetics [13]. The allelic frequencies of CYP3A4*18 among CML patients in Malaysia was almost double (4.44%) that reported among Malaysian diabetics indicating that the polymorphism may play a role in contributing to the disease. However, in another study among the Indian population to associate CYP3A4*18 variant alleles with IM levels, although high inter-patient variability of IM levels were seen, no CYP3A4*18 variants were detected to establish any significant correlation [16].
For CYP3A5, the mutant allele CYP3A5*3 has been reported to contribute to the variable expression in the human liver. This mutation in intron 3, which creates a cryptic splice site, has been reported to cause a premature stop codon, thus resulting in the absence of the CYP3A5 protein [17]. In CYP3A5*3, a guanine (G) replaces an adenine (A) at position 6986. The CYP3A5*1 allele appears to be the main allele associated with CYP3A5 expression and activity. Individuals with at least one CYP3A5*1 polymorphic allele tend to express higher amounts of CYP3A5. In the current study, CML patients who are carriers of the heterozygous (*1/*3) and homozygous variant (*3/*3) genotype of CYP3A5*3 were associated with a significantly lower risk of acquiring resistance against IM, indicating that this variant allele has a protective effect against the development of IM resistance.
In the study conducted by Kim et al. [18] in a Canadian population, the CYP3A5*1/*1 genotype had an adverse impact on achievement of a major cytogenetic response and complete cytogenetic response. Our findings, which indicate that CML patients who are carriers of CYP3A5*1/*1 genotype tend to have a higher risk of developing resistance to IM, are in agreement with those of Kim et al. [18]. On the contrary, a study by Green et al. [19] on 14 Caucasian CML patients on IM therapy demonstrated that CML patients with high CYP3A activity tend to respond better to IM therapy than patients with low activity, indicating that the response may be variable from population to population based on the genetic background of the patients.
CYP3A5*1/*1 genotype was reported to be associated with IM efficacy and *3/*3 genotype with inferior outcome among Egyptian CML patients [20] and also among Indian CML patients [21]. However, Takahashi et al. [22] on Japanese patients and Angelini et al. [23] on Caucasian patients reported no significant association of the CYP3A5*3 polymorphism with IM response. Our findings showed that there is a significant association of CYP3A5*3 polymorphism with IM response but with lower risk for development of resistance. Moreover, we also found that the CYP3A5*1 genotype was higher in the IM-resistant group compared with the IM good-response group. Hence, it is reasonable to suggest that carriers of CYP3A5*3 tend to have a protective effect against acquiring resistance toward IM therapy.
CYP3A5 contributes substantially to the total metabolic clearance of many CYP3A substrates. Since individuals with at least one CYP3A5*1 allele polymorphism express high amounts of CYP3A5, it is reasonable to predict that individuals with the highest clearance and lowest oral bioavailability of CYP3A substrates will be heterozygous or homozygous for CYP3A5*1. Hence, carriers of heterozygous and homozygous CYP3A5*1 genotypes may be more likely to encounter a lack of efficacy from a standard dose of active parent drug. Likewise, the homozygous CYP3A5*3 genotype can lead to a decrease in enzyme activity resulting in lowest clearance and high bioavailability of the drug. Such CML patients are expected to have a better response to IM. As evidenced from our study, CML patients who are carriers of the CYP3A5*1 genotype tend to acquire resistance toward IM treatment, and those with the CYP3A5*3 genotype respond better to IM treatment.
The present study has some limitations in terms of sample size. Further studies are required to acquire a larger patient cohort for long-term IM response monitoring. It would also be worthwhile to estimate the intracellular and plasma levels of both IM and its active metabolites and to correlate their concentrations with the genotype pattern of CYP3A4 and CYP3A5.
Conclusion
Polymorphisms of CYP3A4*18 are rather common among Malaysian CML patients, although they are not significantly associated with response to IM therapy. On the other hand, polymorphism of CYP3A5*3 is associated with IM treatment response in Malaysian CML patients with individuals having the CYP3A5*1/*3 and CYP3A5*3/*3 genotypes posing lower risks of being resistant to IM treatment. Therefore, pretreatment genotyping of this SNP may be important in predicting IM response in CML patients.
References
Moen MD, McKeage K, Plosker GL, Siddiqui MAA. Imatinib—a review of its use in chronic myeloid leukaemia. Drugs. 2007;67(2):299–320. doi:10.2165/00003495-200767020-00010.
Au A, Aziz Baba A, Goh AS, Wahid Fadilah SA, Teh A, Rosline H, Ankathil R. Association of genotypes and haplotypes of multi-drug transporter genes ABCB1 and ABCG2 with clinical response to imatinib mesylate in chronic myeloid leukemia patients. Biomed Pharmacother. 2014;68(3):343–9. doi:10.1016/j.biopha.2014.01.009.
Bajpai P, Tripathi AK, Agrawal D. Genetic polymorphism of CYP3A5 in Indian chronic myeloid leukemia patients. Mol Cell Biochem. 2010;336(1–2):49–54. doi:10.1007/s11010-009-0268-1.
Ota T, Kamada Y, Hayashida M, Iwao-Koizumi K, Murata S, Kinoshita K. Combination analysis in genetic polymorphisms of drug-metabolizing enzymes CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A5 in the Japanese population. Int J Med Sci. 2015;12(1):78–82. doi:10.7150/ijms.10263.
Peng B, Dutreix C, Mehring G, Hayes MJ, Ben-Am M, Seiberling M, Pokorny R, Capdeville R, Lloyd P. Absolute bioavailability of imatinib (Glivec) orally versus intravenous infusion. J Clin Pharmacol. 2004;44(2):158–62. doi:10.1177/0091270003262101.
Hu YF, He J, Chen GL, Wang D, Liu ZQ, Zhang C, Duan LF, Zhou HH. CYP3A5*3 and CYP3A4*18 single nucleotide polymorphisms in a Chinese population. Clin Chimi Acta. 2005;353(1–2):187–92. doi:10.1016/j.cccn.2004.11.005.
Seong SJ, Lim M, Sohn SK, Moon JH, Oh SJ, Kim BS, Ryoo HM, Chung JS, Joo YD, Bang SM, Jung CW, Kim DH, Park SY, Yoon SS, Kim I, Lee HG, Won JH, Min YH, Cheong JW, Park JS, Eom KS, Hyun MS, Kim MK, Kim H, Park MR, Park J, Kim CS, Kim HJ, Kim YK, Park EK, Zang DY, Jo DY, Lee HW, Yoon YR. Influence of enzyme and transporter polymorphisms on trough imatinib concentration and clinical response in chronic myeloid leukemia patients. Ann Oncol. 2013;24(3):756–60. doi:10.1093/annonc/mds532.
Dai D, Tang J, Rose R, Hodgson E, Bienstock RJ, Mohrenweiser HW, Goldstein JA. Identification of variants of CYP3A4 and characterization of their abilities to metabolize testosterone and chlorpyrifos. J Pharmacol Exp Ther. 2001;299(3):825–31.
Park J-Y, Cha Y-J, Kim K-A. CYP3A5*3 polymorphism and its clinical implications and pharmacokinetic role. Transl Clin Pharmacol. 2014;22(1):3–7.
Baccarani M, Deininger MW, Rosti G, Hochhaus A, Soverini S, Apperley JF, Cervantes F, Clark RE, Cortes JE, Guilhot F, Hjorth-Hansen H, Hughes TP, Kantarjian HM, Kim DW, Larson RA, Lipton JH, Mahon FX, Martinelli G, Mayer J, Muller MC, Niederwieser D, Pane F, Radich JP, Rousselot P, Saglio G, Saussele S, Schiffer C, Silver R, Simonsson B, Steegmann JL, Goldman JM, Hehlmann R. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood. 2013;122(6):872–84. doi:10.1182/blood-2013-05-501569.
Baccarani M, Pileri S, Steegmann JL, Muller M, Soverini S, Dreyling M. Chronic myeloid leukemia: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23(7):72–7. doi:10.1093/annonc/mds228.
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi:10.1093/bioinformatics/bth457.
Ruzilawati AB, Suhaimi AW, Gan SH. Genetic polymorphisms of CYP3A4: CYP3A4*18 allele is found in five healthy Malaysian subjects. Clin Chim Acta. 2007;383(1–2):158–62. doi:10.1016/j.cca.2007.05.004.
Lee SJ, Lee SS, Jeong HE, Shon JH, Ryu JY, Sunwoo YE, Liu KH, Kang W, Park YJ, Shin CM, Shin JG. The CYP3A4*18 allele, the most frequent coding variant in Asian populations, does not significantly affect the midazolam disposition in heterozygous individuals. Drug Metab Dispos. 2007;35(11):2095–101. doi:10.1124/dmd.107.016733.
Yamamoto T, Nagafuchi N, Ozeki T, Kubota T, Ishikawa H, Ogawa S, Yamada Y, Hirai H, Iga T. CYP3A4*18: it is not rare allele in Japanese population. Drug Metab Pharmacokinet. 2003;18(4):267–8.
Gota V, Sharma A, Paradkar A, Patil A, Khattry N, Menon H, Choughule A, Amare P, Banavali S, Arora B. Association between CYP3A4 polymorphisms, trough imatinib plasma concentration and cytogenetic response in chronic phase chronic myeloid leukemia (CML-CP). Blood. 2012;120(21):3788.
Lee SJ, Goldstein JA. Functionally defective or altered CYP3A4 and CYP3A5 single nucleotide polymorphisms and their detection with genotyping tests. Pharmacogenomics. 2005;6(4):357–71. doi:10.1517/14622416.6.4.357.
Kim DH, Sriharsha L, Xu W, Kamel-Reid S, Liu X, Siminovitch K, Messner HA, Lipton JH. Clinical relevance of a pharmacogenetic approach using multiple candidate genes to predict response and resistance to imatinib therapy in chronic myeloid leukemia. Clin Cancer Res. 2009;15(14):4750–8. doi:10.1158/1078-0432.ccr-09-0145.
Green H, Skoglund K, Rommel F, Mirghani RA, Lotfi K. CYP3A activity influences imatinib response in patients with chronic myeloid leukemia: a pilot study on in vivo CYP3A activity. Eur J Clin Pharmacol. 2010;66(4):383–6. doi:10.1007/s00228-009-0772-y.
Bedewy AM, El-Maghraby SM. Do SLCO1B3 (T334G) and CYP3A5*3 polymorphisms affect response in Egyptian chronic myeloid leukemia patients receiving imatinib therapy? Hematology. 2013;18(4):211–6. doi:10.1179/1607845412Y.0000000067.
Sailaja K, Rao DN, Rao DR, Vishnupriya S. Analysis of CYP3A5*3 and CYP3A5*6 gene polymorphisms in Indian chronic myeloid leukemia patients. Asian Pac J Cancer Prev. 2010;11(3):781–4.
Takahashi N, Miura M, Scott SA, Kagaya H, Kameoka Y, Tagawa H, Saitoh H, Fujishima N, Yoshioka T, Hirokawa M, Sawada K. Influence of CYP3A5 and drug transporter polymorphisms on imatinib trough concentration and clinical response among patients with chronic phase chronic myeloid leukemia. J Hum Genet. 2010;55(11):731–7. doi:10.1038/jhg.2010.98.
Angelini S, Soverini S, Ravegnini G, Barnett M, Turrini E, Thornquist M, Pane F, Hughes TP, White DL, Radich J, Kim DW, Saglio G, Cilloni D, Iacobucci I, Perini G, Woodman R, Cantelli-Forti G, Baccarani M, Hrelia P, Martinelli G. Association between imatinib transporters and metabolizing enzymes genotype and response in newly diagnosed chronic myeloid leukemia patients receiving imatinib therapy. Haematologica. 2013;98(2):193–200. doi:10.3324/haematol.2012.066480.
Acknowledgements
This study was supported by Universiti Sains Malaysia Research grant no. 1001/PPSP/812103. All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this manuscript, take responsibility for the integrity of the work as a whole and have given final approval for the version to be published.
Disclosures
Najlaa Maddin, Azlan Husin, Siew Hua Gan, Baba Abdul Aziz and Ravindran Ankathil declare no conflict of interest.
Compliance with Ethics Guidelines
All procedures performed in the studies involving human participants were in accordance with ethical standards of the Research and Ethic Committee of Universiti Sains Malaysia and Ministry of Health, Malaysia, and with the Declaration of Helsinki of 1964, as revised in 2013. Informed consent was obtained from all patients for being included in the study.
Data Availability
All data generated or analyzed during this study are included in this published article.
Open Access
This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Author information
Authors and Affiliations
Corresponding author
Additional information
Enhanced Content
To view enhanced content for this article go to http://www.medengine.com/Redeem/3227F06024D4129C.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/licenses/by/4.0), which permits use, duplication, adaptation, distribution, and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Maddin, N., Husin, A., Gan, S.H. et al. Impact of CYP3A4*18 and CYP3A5*3 Polymorphisms on Imatinib Mesylate Response Among Chronic Myeloid Leukemia Patients in Malaysia. Oncol Ther 4, 303–314 (2016). https://doi.org/10.1007/s40487-016-0035-x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40487-016-0035-x