Abstract
Objective
The aims of this study were to determine the effects of the CYP2C8*3 and *4 polymorphisms on imatinib metabolism and plasma imatinib concentrations in chronic myeloid leukaemia (CML) patients.
Methods
We genotyped 210 CML patients from the TIDELII trial receiving imatinib 400–800 mg/day for CYP2C8*3 (rs11572080, rs10509681) and *4 (rs1058930). Steady-state trough total plasma N-desmethyl imatinib (major metabolite):imatinib concentration ratios (metabolic ratios) and trough total plasma imatinib concentrations were compared between genotypes (one-way ANOVA with Tukey post hoc).
Results
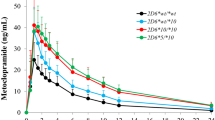
CYP2C8*3 (n = 34) and *4 (n = 15) carriers had significantly higher (P < 0.01) and lower (P < 0.01) metabolic ratios, respectively, than CYP2C8*1/*1 (n = 147) patients (median ± standard deviation: 0.28 ± 0.08, 0.18 ± 0.06 and 0.22 ± 0.08, respectively). Plasma imatinib concentrations were consequently > 50% higher for CYP2C8*1/*4 than for CYP2C8*1/*1 and CYP2C8*3 carriers (2.18 ± 0.66 vs. 1.45 ± 0.74 [P < 0.05] and 1.36 ± 0.98 μg/mL [P < 0.05], respectively).
Conclusions
CYP2C8 genotype significantly alters imatinib metabolism in patients through gain- and loss-of-function mechanisms.



Similar content being viewed by others
References
Maino E, Sancetta R, Viero P, Imbergamo S, Scattolin AM, Vespignani M, et al. Current and future management of Ph/BCR-ABL positive ALL. Expert Rev Anticancer Ther. 2014;14(6):723–40. doi:10.1586/14737140.2014.895669.
Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2014 update on diagnosis, monitoring, and management. Am J Hematol. 2014;89(5):547–56. doi:10.1002/ajh.23691.
Linch M, Claus J, Benson C. Update on imatinib for gastrointestinal stromal tumors: duration of treatment. Onco Targets Ther. 2013;6:1011–23. doi:10.2147/ott.s31260.
Gleevec NDA 021588 label information. Revised Sep 2016. US Food and Drug Administration, Center for Drug Evaluation and Research; 2016. http://www.accessdata.fda.gov/drugsatfda_docs/label/2016/021588s047lbl.pdf. Accessed 13 Oct 2016.
Gotta V, Bouchet S, Widmer N, Schuld P, Decosterd LA, Buclin T, et al. Large-scale imatinib dose-concentration-effect study in CML patients under routine care conditions. Leuk Res. 2014;38(7):764–72. doi:10.1016/j.leukres.2014.03.023.
Widmer N, Bardin C, Chatelut E, Paci A, Beijnen J, Leveque D, et al. Review of therapeutic drug monitoring of anticancer drugs part two-targeted therapies. Eur J Cancer. 2014;50(12):2020–36. doi:10.1016/j.ejca.2014.04.015.
Picard S, Titier K, Etienne G, Teilhet E, Ducint D, Bernard MA, et al. Trough imatinib plasma levels are associated with both cytogenetic and molecular responses to standard-dose imatinib in chronic myeloid leukemia. Blood. 2007;109(8):3496–9. doi:10.1182/blood-2006-07-036012.
Larson RA, Druker BJ, Guilhot F, O’Brien SG, Riviere GJ, Krahnke T, et al. Imatinib pharmacokinetics and its correlation with response and safety in chronic-phase chronic myeloid leukemia. Blood. 2008;111(8):4022–8. doi:10.1182/blood-2007-10-116475.
Berglund E, Ubhayasekera SJ, Karlsson F, Akcakaya P, Aluthgedara W, Ahlen J, et al. Intracellular concentration of the tyrosine kinase inhibitor imatinib in gastrointestinal stromal tumor cells. Anticancer Drugs. 2014;25(4):415–22. doi:10.1097/cad.0000000000000069.
Mlejnek P, Dolezel P, Faber E, Kosztyu P. Interactions of N-desmethyl imatinib, an active metabolite of imatinib, with P-glycoprotein in human leukemia cells. Ann Hematol. 2011;90(7):837–42. doi:10.1007/s00277-010-1142-7.
Manley PW, Blasco F, Mestan J, Aichholz R. The kinetic deuterium isotope effect as applied to metabolic deactivation of imatinib to the des-methyl metabolite, CGP74588. Bioorg Med Chem. 2013;21(11):3231–9. doi:10.1016/j.bmc.2013.03.038.
Skoglund K, Boiso Moreno S, Jonsson JI, Vikingsson S, Carlsson B, Green H. Single-nucleotide polymorphisms of ABCG2 increase the efficacy of tyrosine kinase inhibitors in the K562 chronic myeloid leukemia cell line. Pharmacogenet Genomics. 2014;24(1):52–61. doi:10.1097/FPC.0000000000000022.
Skoglund K, Moreno SB, Baytar M, Jonsson JI, Green H. ABCB1 haplotypes do not influence transport or efficacy of tyrosine kinase inhibitors in vitro. Pharmgenomics Pers Med. 2013;6:63–72. doi:10.2147/PGPM.S45522.
Josephs DH, Fisher DS, Spicer J, Flanagan RJ. Clinical pharmacokinetics of tyrosine kinase inhibitors: implications for therapeutic drug monitoring. Ther Drug Monit. 2013;35(5):562–87. doi:10.1097/FTD.0b013e318292b931.
Gschwind HP, Pfaar U, Waldmeier F, Zollinger M, Sayer C, Zbinden P, et al. Metabolism and disposition of imatinib mesylate in healthy volunteers. Drug Metab Dispos. 2005;33(10):1503–12. doi:10.1124/dmd.105.004283.
Di Gion P, Kanefendt F, Lindauer A, Scheffler M, Doroshyenko O, Fuhr U, et al. Clinical pharmacokinetics of tyrosine kinase inhibitors: focus on pyrimidines, pyridines and pyrroles. Clin Pharmacokinet. 2011;50(9):551–603. doi:10.2165/11593320-000000000-00000.
Bouchet S, Titier K, Moore N, Lassalle R, Ambrosino B, Poulette S, et al. Therapeutic drug monitoring of imatinib in chronic myeloid leukemia: experience from 1216 patients at a centralized laboratory. Fundam Clin Pharmacol. 2013;27(6):690–7. doi:10.1111/fcp.12007.
de Wit D, Guchelaar HJ, den Hartigh J, Gelderblom H, van Erp NP. Individualized dosing of tyrosine kinase inhibitors: are we there yet? Drug Discov Today. 2015;20(1):18–36. doi:10.1016/j.drudis.2014.09.007.
Filppula AM, Laitila J, Neuvonen PJ, Backman JT. Potent mechanism-based inhibition of CYP3A4 by imatinib explains its liability to interact with CYP3A4 substrates. Br J Pharmacol. 2012;165(8):2787–98. doi:10.1111/j.1476-5381.2011.01732.x.
Filppula AM, Neuvonen M, Laitila J, Neuvonen PJ, Backman JT. Autoinhibition of CYP3A4 leads to important role of CYP2C8 in imatinib metabolism. Drug Metab Dispos. 2013;41(1):50–9. doi:10.1124/dmd.112.048017.
Nebot N, Crettol S, d’Esposito F, Tattam B, Hibbs DE, Murray M. Participation of CYP2C8 and CYP3A4 in the N-demethylation of imatinib in human hepatic microsomes. Br J Pharmacol. 2010;161(5):1059–69. doi:10.1111/j.1476-5381.2010.00946.x.
Skoglund K, Richter J, Olsson-Stromberg U, Bergquist J, Aluthgedara W, Ubhayasekera SJ, et al. In vivo cytochrome P450 3A isoenzyme activity and pharmacokinetics of imatinib in relation to therapeutic outcome in patients with chronic myeloid leukemia. Ther Drug Monit. 2016;38(2):230–8. doi:10.1097/FTD.0000000000000268.
Gurney H, Wong M, Balleine RL, Rivory LP, McLachlan AJ, Hoskins JM, et al. Imatinib disposition and ABCB1 (MDR1, P-glycoprotein) genotype. Clin Pharmacol Ther. 2007;82(1):33–40. doi:10.1038/sj.clpt.6100201.
Widmer N, Decosterd LA, Csajka C, Leyvraz S, Duchosal MA, Rosselet A, et al. Population pharmacokinetics of imatinib and the role of alpha-acid glycoprotein [published erratum appears in Br J Clin Pharmacol 2010; 70 (2):316]. Br J Clin Pharmacol. 2006;62(1):97–112. doi:10.1111/j.1365-2125.2006.02719.x.
Schmidli H, Peng B, Riviere GJ, Capdeville R, Hensley M, Gathmann I, et al. Population pharmacokinetics of imatinib mesylate in patients with chronic-phase chronic myeloid leukaemia: results of a phase III study. Br J Clin Pharmacol. 2005;60(1):35–44. doi:10.1111/j.1365-2125.2005.02372.x.
van Erp NP, Gelderblom H, Karlsson MO, Li J, Zhao M, Ouwerkerk J, et al. Influence of CYP3A4 inhibition on the steady-state pharmacokinetics of imatinib. Clin Cancer Res. 2007;13(24):7394–400. doi:10.1158/1078-0432.CCR-07-0346.
Singh O, Chan JY, Lin K, Heng CC, Chowbay B. SLC22A1-ABCB1 haplotype profiles predict imatinib pharmacokinetics in asian patients with chronic myeloid leukemia. PLoS One. 2012;7(12):e51771. doi:10.1371/journal.pone.0051771.
Seong SJ, Lim M, Sohn SK, Moon JH, Oh SJ, Kim BS, et al. Influence of enzyme and transporter polymorphisms on trough imatinib concentration and clinical response in chronic myeloid leukemia patients. Ann Oncol. 2013;24(3):756–60. doi:10.1093/annonc/mds532.
Yamakawa Y, Hamada A, Nakashima R, Yuki M, Hirayama C, Kawaguchi T, et al. Association of genetic polymorphisms in the influx transporter SLCO1B3 and the efflux transporter ABCB1 with imatinib pharmacokinetics in patients with chronic myeloid leukemia. Ther Drug Monit. 2011;33(2):244–50. doi:10.1097/FTD.0b013e31820beb02.
Takahashi N, Miura M, Scott SA, Kagaya H, Kameoka Y, Tagawa H, et al. Influence of CYP3A5 and drug transporter polymorphisms on imatinib trough concentration and clinical response among patients with chronic phase chronic myeloid leukemia. J Hum Genet. 2010;55(11):731–7. doi:10.1038/jhg.2010.98.
Khan MS, Barratt DT, Somogyi AA. Impact of CYP2C8*3 polymorphism on in vitro metabolism of imatinib to N-desmethyl imatinib. Xenobiotica. 2015:1–10. doi: 10.3109/00498254.2015.1060649.
Bahadur N, Leathart JB, Mutch E, Steimel-Crespi D, Dunn SA, Gilissen R, et al. CYP2C8 polymorphisms in Caucasians and their relationship with paclitaxel 6alpha-hydroxylase activity in human liver microsomes. Biochem Pharmacol. 2002;64(11):1579–89.
Gao Y, Liu D, Wang H, Zhu J, Chen C. Functional characterization of five CYP2C8 variants and prediction of CYP2C8 genotype-dependent effects on in vitro and in vivo drug-drug interactions. Xenobiotica. 2010;40(7):467–75. doi:10.3109/00498254.2010.487163.
Jiang H, Zhong F, Sun L, Feng W, Huang ZX, Tan X. Structural and functional insights into polymorphic enzymes of cytochrome P450 2C8. Amino Acids. 2011;40(4):1195–204. doi:10.1007/s00726-010-0743-8.
Wang Z, Wang S, Huang M, Hu H, Yu L, Zeng S. Characterizing the effect of cytochrome P450 (CYP) 2C8, CYP2C9, and CYP2D6 genetic polymorphisms on stereoselective N-demethylation of fluoxetine. Chirality. 2014;26(3):166–73. doi:10.1002/chir.22289.
Rochat B, Zoete V, Grosdidier A, von Grunigen S, Marull M, Michielin O. In vitro biotransformation of imatinib by the tumor expressed CYP1A1 and CYP1B1. Biopharm Drug Dispos. 2008;29(2):103–18. doi:10.1002/bdd.598.
Yeung DT, Osborn MP, White DL, Branford S, Braley J, Herschtal A, et al. TIDEL-II: first-line use of imatinib in CML with early switch to nilotinib for failure to achieve time-dependent molecular targets. Blood. 2015;125(6):915–23. doi:10.1182/blood-2014-07-590315.
R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013.
Fox J, Weisberg S. An R companion to applied regression. 2nd ed. Thousand Oaks: Sage; 2011.
Hothorn T, Bretz F, Westfall P. Simultaneous inference in general parametric models. Biom J. 2008;50(3):346–63.
Bates D, Maechler M, Bolker B, Wlaker S. lme4: linear mixed-effects models using Eigen and S4. R package version 1.0-5. 2013. http://CRAN.R-project.org/package=lme4. Accessed 19 Apr 2016.
Fox J. Effect displays in R for generalised linear models. J Stat Softw. 2003;8(15):1–27.
Shimada T, Yamazaki H, Mimura M, Inui Y, Guengerich FP. Interindividual variations in human liver cytochrome P-450 enzymes involved in the oxidation of drugs, carcinogens and toxic chemicals: studies with liver microsomes of 30 Japanese and 30 Caucasians. J Pharmacol Exp Ther. 1994;270(1):414–23.
Di Paolo A, Polillo M, Capecchi M, Cervetti G, Barate C, Angelini S, et al. The c.480C > G polymorphism of hOCT1 influences imatinib clearance in patients affected by chronic myeloid leukemia. Pharmacogenomics J. 2014;14(4):328–35. doi:10.1038/tpj.2014.7.
Gandia P, Arellano C, Lafont T, Huguet F, Malard L, Chatelut E. Should therapeutic drug monitoring of the unbound fraction of imatinib and its main active metabolite N-desmethyl-imatinib be developed? Cancer Chemother Pharmacol. 2013;71:531–6. doi:10.1007/s00280-012-2035-3.
Gater A, Heron L, Abetz-Webb L, Coombs J, Simmons J, Guilhot F, et al. Adherence to oral tyrosine kinase inhibitor therapies in chronic myeloid leukemia. Leuk Res. 2012;36(7):817–25. doi:10.1016/j.leukres.2012.01.021.
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi:10.1038/nature11632.
Stingl Kirchheiner JC, Brockmoller J. Why, when, and how should pharmacogenetics be applied in clinical studies? Current and future approaches to study designs. Clin Pharmacol Ther. 2011;89(2):198–209. doi:10.1038/clpt.2010.274.
Ritchie MD. The success of pharmacogenomics in moving genetic association studies from bench to bedside: study design and implementation of precision medicine in the post-GWAS era. Hum Genet. 2012;131(10):1615–26. doi:10.1007/s00439-012-1221-z.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This study was funded by the School of Medical Sciences (now Adelaide Medical School), University of Adelaide and a Royal Adelaide Hospital Research Foundation Clinical Project Grant.
Conflicts of Interest
DLW received research funding and honoraria from Novartis and BMS. TPH acted on the advisory board of, and received research funding and honoraria from, Novartis, BMS and Ariad. DTY received research funding and honoraria from, Novartis, BMS and Ariad. DTB, HKC, AM and AAS have no conflicts of interest to declare.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Barratt, D.T., Cox, H.K., Menelaou, A. et al. CYP2C8 Genotype Significantly Alters Imatinib Metabolism in Chronic Myeloid Leukaemia Patients. Clin Pharmacokinet 56, 977–985 (2017). https://doi.org/10.1007/s40262-016-0494-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40262-016-0494-0