Abstract
The well-known SIR epidemic model is revisited. Continuous-time and discrete-time versions of an alternative model of this class are presented, discussed and validated with actual data. The proposed model follows from the calculation of the mean number of new infected cases due to the eventual meeting of susceptible and infected individuals, based on a simple probabilistic argument. Determination of the invariant set in the state space and convergence conditions towards equilibrium are established. For numerical analysis, data of daily number of new diagnosed cases provided by the Brazilian Ministry of Health and World Health Organization of COVID-19 outbreak that currently occurs respectively in Brazil and in the UK are used. Illustrations and model prediction analysis are provided and discussed from full data of both aforementioned countries which include more than 400 epidemic days. Three different and complementary strategies for parameter identification including the impact of causality on the optimal solution of the nonlinear mean square problem are discussed.
Similar content being viewed by others
1 Introduction
We are living in a new time characterized by an unprecedented demand of the health system. In Brazil, we have the Unified Health System (SUS) that is showing its importance to better serve the population of our country. It is necessary to highlight the commitment and dedication of all health professionals and a significant part of the population, which also deserves praise, as they seek to maintain an effective social distance, even in the face of unreasonable opinions that, unlike most of the world, insist to minimize its beneficial effects. From the public health point of view, it is necessary to be able to evaluate possible scenarios and validating actions in order to flatten the peak of the epidemic and preserve the hospital’s service capacity. Together with vaccination, social distancing (including the use of masks) is perhaps one of the most effective actions to be implemented, but the key question is how to assess its effectiveness and how to decide when and how to mitigate it, without allowing new waves of the epidemic. The research effort on mathematical models development appears to be a possible way to find an adequate answer to this concern, since a sufficiently precise model would be an appropriate device to predict and quantify the epidemic time evolution.
This paper is entirely devoted to the study of the most well known class of epidemic models known by the acronym SIR, which stands for Susceptible–Infected–Removed, formulated in both continuous-time and discrete-time domains. Nowadays, the literature presents countless studies dealing with epidemics. Among them, it is important to put in evidence the deterministic continuous-time modeling presented in the seminal paper by Kermack and McKendrick (1927), almost a century ago, which established a solid mathematical basis for further developments.
Since then, this research area has been developed by incorporating to the original class many other subclasses like SEIR, SEIRS, SIRS, SEI, SEIS, SI, SIS, among others. The reader is asked to see the survey paper by Hethcote (2000) where this subject is deeply treated. In addition, Hethcote (2000) provided a rather complete set of results and references putting in clear evidence the more representative contributions made over time. Furthermore, it is important to mention the books of Anderson (1987), Bailey (1975) and Brauer and Castillo-Chavez (2012) including the references therein as excellent sources of information on model development analysis, control design and many other related topics on infectious diseases.
All mentioned subclasses of the SIR model are readily recognized by the infection mechanism which includes into the model a nonlinear term expressed, putting aside normalization, by a product of variables, see Kermack and McKendrick (1927)), Soares (2011) and the very recent papers by Giordano et al. (2020) and Silva et al. (2020). Here, we intend to go further by proposing a new approach to determine the spread of the infection in a population of given constant size. In our opinion, the proposed model is a valid theoretical alternative to the classical SIR model but its final validation needs to be established in practice from actual data.
The literature provides many studies dealing with the continuous-time version of the SIR model but much less about the discrete-time one. For the last mentioned subject, the interested reader is invited to see Anastassopoulou et al. (2020), Brauer and Castillo-Chavez (2012), Calafiore et al. (2020) and Hu et al. (2014) including, in the former reference, a useful treatment of difference equations. The subclasses of the SIR model are all expressed by nonlinear differential or difference equations, in continuous-time or discrete-time domains, respectively. The order of the model depends on the number of state variables needed to discriminate the various types of individuals in the population. Recently, in Giordano et al. (2020), a new class of SIR model expressed in terms of a eighth-order differential equation has been proposed with the main goal to evaluate possible scenarios of the COVID-19 outbreak evolution in Italy.
To be as faithful as possible in the face of reality, models of the SIR class must be considered time-varying because it is necessary to allow its parameters to vary over time in order to capture trends in how the population behaves during the outbreak time evolution. This makes the parameter identification step much more complicated when compared to the time invariant case. In particular, a sort of causality property arises that by consequence must be imposed such that past optimal values of the estimated parameters are not affected whenever new measurements are incorporated into the data set. This aspect is important since causality is a suitable property to be present in any dynamic model. Hence, in this context, three parameter estimation strategies are presented and discussed. The discrete-time versions of the classical model and the proposed model of the SIR class are validated using daily data of the COVID-19 outbreak presently occurring in Brazil and in the UK, a country where the epidemic is approaching the end. In both countries, the results put in clear evidence the existence of successive epidemic waves. Moreover, since new data are provided daily, it seems more natural to apply the discrete-time versions of the models and identify the time-varying parameters. To this end, data from more than 400 days of COVID-19 outbreaks occurring in Brazil and in the UK are used to put in evidence new modelling aspects including robustness with respect to data errors.
This is an extended version of the paper Costa et al. (2020) by the same authors. Besides the material that appeared in that reference, this paper incorporates the development and analysis of the continuous-time version of the proposed model of the SIR class and uses the full data set available until now (more than 400 days) for model identification and validation. Instead of Italy, data from the United Kingdom have been considered because, as already mentioned, in this country the outbreak is now approaching the end.
The paper is organized as follows: In the next section, the classical model of the SIR class in continuous time is presented. In Sect. 3, an alternative model of the SIR class is proposed and discussed with special attention to the mechanism of infection that is primarily responsible for the spread of the disease in the population. In the same section, a unified model in continuous time is constructed and the relationship between both models is established. Section 4 is entirely devoted to develop the unified model in discrete time, putting in evidence the stability properties and three strategies for parameter identification. In Sect. 5, the results are applied to the outbreak that is taking place in Brazil and the one that occurred in the UK. The main conclusions and recommendations are summarized in Sect. 6.
The notation used throughout is standard. Specifically, the symbols \(\mathbb {R}\), \(\mathbb {R}_+\), and \(\mathbb {N}\) denote the sets of real, real non-negative, and natural numbers, respectively. A function or trajectory f(t) evaluated at some time instant \(t=t_k \in \mathbb {R}\) is denoted as \(f[k]=f(t_k)\) for all \(k \in \mathbb {N}\).
2 The Classical SIR Model
The classical deterministic Susceptible–Infected–Removed (SIR) model was proposed for the first time almost one century ago in the seminal paper by Kermack and McKendrick (1927).Footnote 1 We now discuss this model as originally formulated in continuous time but in a probabilistic context that makes possible some interpretations and comparisons in the unified viewpoint to be given afterwards; see also Silva et al. (2020).
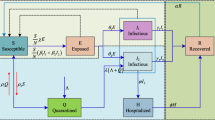
The independent variable \(t \in \mathbb {R}_+\) denotes time and the initial time \(t_0=0\) corresponds to the time instant on which the first case of infection was diagnosed. The population \(\mathcal{P}\) made up of M individuals is split into three types, and at each time instant each person is supposed to belong to only one of them, namely:
-
Susceptible (S)—is the group of healthy individuals. The total number of elements in this set, denoted by s(t), indicates the number of healthy individuals on \(t \in \mathbb {R}_+\).
-
Infected (I)—is the group of infected individuals. The total number of elements in this set, denoted by i(t), indicates the number of infected individuals, capable of transmitting the disease, on \(t \in \mathbb {R}_+\).
-
Removed (R)—is the group of individuals who no longer have the ability to transmit the disease because they are immunized or dead.Footnote 2 The total number of elements in this set is denoted by r(t), on time \(t \in \mathbb {R}_+\).
By assumption, the population remains constant throughout the epidemic horizon, births are not taken into account, which implies \(s(t) + i(t) + r(t) = M\) for all \(t \in \mathbb {R}_+\). Let x(t) be any member of the population on time \(t \in \mathbb {R}_+\). Without any further information, at time \(t \in \mathbb {R}_+\), the probability to be of type susceptible, infected, or removed is s(t)/M, i(t)/M or r(t)/M, respectively. The key issue and the main characteristic of any model of the SIR class is the mechanism that determines the susceptible to infected transition which is responsible for its nonlinear nature. In the mathematical framework of SIR models, the ones of interest are developed and interpreted in the sequel.
2.1 The Classical SIR Model
Following Hethcote (2000), consider the time-varying version of the classical SIR model with probabilistic interpretation. To this end, let \(\beta (t)\) be the average number of effective contacts, those which result in infection of a person per unit of time. Hence, \(\beta (t) (i(t)/M)\) is the average number of effective contacts with infected of one individual of the population per unit of time. Taking into account that at time \(t \in \mathbb {R}_+\) the number of susceptible is s(t), then the average number of new infected in an arbitrarily small time interval \(\Delta t >0\) is
In the aforementioned reference the time-invariant case \(\beta (t) = \beta \) is considered. Moreover, by comparison with the model resulting from the mass action law, the parameter dependence \(\beta = \eta M^\upsilon \) for some \((\eta , \upsilon )\) is discussed. Measurements strongly suggest that \(\upsilon \approx 0\), see Hethcote (2000) for more details on this relevant aspect.
3 The Proposed SIR Model
We now propose an alternative model of the SIR class. To this end, let us consider the following experiment. In an arbitrary instant of time \(t \in \mathbb {R}_+\), let a pair of individuals \((x_1, x_2)\) from the population \(\mathcal{P}\) be randomly chosen, with replacement.Footnote 3 The probability that \(x_1\) is healthy (\(x_1 \in S\)) and \(x_2\) is infected (\(x_2 \in I\)), or vice versa, is 2(s(t)/M)(i(t)/M). Assuming that, a fraction p(t) of healthy people becomes infected due to the meeting of healthy and infected individuals, per unit of time, then the average number of new people infected in an arbitrarily small time interval \(\Delta t > 0\) is given by
where \(\gamma (t) = 2 p(t) \in (0, 2)\). Note that the first term s(t) in the product shown in the first equality of (2), indicates that only healthy people, when they meet infected people, can become infected. At this point, it is interesting to compare the estimated number of new infected people provided by both SIR models. From (1) and (2), it follows that
from which some conclusions can be drawn. First, at the very beginning of the epidemic evolution, the fact that \(s(t) \approx M\) implies \(\beta (t) \approx \gamma (t)\), meaning that both models virtually coincide. Of course, the same fact does not remain true whenever the epidemic evolves in time and the number of susceptible persons becomes smaller. This fact can be verified by comparing the phase-plane drawing of both models. Second, even though \(\beta (t)\) may not depend on the population size M, it may be time-varying and be linearly dependent on the density of susceptible in the population as indicated in (3). Indeed, among many factors, it may depend on the behavior changes, at least in part of the population, due to alerts and awareness campaigns.
3.1 The Unified Model in Continuous Time
Since the infection mechanism has already been determined, the time evolution of the state variables of both models (s(t), i(t), r(t)) for all \(t \in \mathbb {R}_+\), can be written in a unified manner. To this end, let us define the selection index
and perform the limit \(\Delta t \rightarrow 0\) to get the following state space equations of a nonlinear time-varying dynamic system that can be expressed in the form
where \(\gamma (t) \in (0, 2)\), \(\alpha (t) > 0\), for all \(t \in \mathbb {R}_+\) and non-negative initial conditions \(s(0)=s_0\), \(i(0)=i_0\) and \(r(0)=r_0\) satisfying \(s_0 + i_0 + r_0 = M\). The parameters \(\alpha = \alpha (t)\) and \(\gamma = \gamma (t)\) are considered time-varying because there is strong evidence that they change in the course of the epidemic evolution, due to the reasons mentioned before. However, in some instances, the parameter \(\alpha = \alpha (t)\) can be considered time-invariant, and determined if we know the half-life of the process that characterizes the transition of infected individuals to removed, under the hypothesis that no contagion occurs. The half-life \(L_h\) expressed in units of time yields
that is, for a half-life of \(L_h= 7\) days, we obtain \(1/\alpha \approx 10\) days, which from actual data seems to be quite reasonable. By its turn, the parameter \(\gamma = \gamma (t)\) expresses the rate at which the infection spreads over time and whenever it decreases, results in a gradual reduction of the number of infected persons. A possible interpretation is that the parameter \(\alpha (t)\) is a characteristic of the disease while \(\gamma (t)\) strongly depends on the population behavior. Certainly, it is a major consequence of the social distancing and additional precautions adopted by the population in some time intervals.
3.2 The Basic Reproduction Number
In epidemiology, there is a number that defines the secondary infections produced by one infected individual being introduced in a susceptible individuals group (Hethcote 2000). This number (which in the present case depends on time) called basic reproduction number, denoted as \(R_0\), in our time-varying SIR models can be calculated as
Hence, from (6), it is clear that for values of \(R_\nu > 1\), the infection spreads in the susceptible population, and on the contrary, whenever \(R_\nu < 1\) the infection declines. The parameter \(R_0\), an upper bound to \(R_\nu \), has a vital role in the study of epidemics, and in the case of our time-varying models, it helps us to observe how the epidemic is evolving in the population and approaches the end since \(R_0 <1\) implies that \(R_\nu <1\). Clearly, \(R_0\) depends only on the model parameters (it does not depend on s(t)) and as it can be verified \(R_0(t) \ge R_1(t) \ge R_2(t)\), for all \(t \in \mathbb {R}_+\).
The proposed SIR model that we have just obtained has an intrinsic hypothesis that seems to be unrealistic. On a time interval, the average number of new infections is given by (2). To obtain this value, we have assumed that each individual in the population can meet any other, with equal probability. We believe that this simplifying hypothesis is no longer realistic when, for example, the population spreads over a large area with a non-uniform demographic density. The impact of this hypothesis, in face of reality, is difficult to measure. In fact, the possibility that all individuals meet each other tends to increase the number of new infected, but not taking into account the eventual existence of high population densities, in some regions, acts in the opposite direction. Fortunately, as we will see later, this undesirable aspect can be mitigated if we consider time-varying models, as in (5)–(7) with the parameters being determined such as the modeling error is minimum. Clearly, the same reasoning is valid for the classical SIR model resulting from \(\nu =1\). Moreover, dividing each equation in (5)–(7) by the population size M, it is simple to see that the property stated in the following remark holds.
Remark 1
The dynamic Eqs. (5)–(7) are such that the normalization \(M=1\) can be imposed, with no loss of generality, for both models \(\nu \in \{1, 2 \}\).
3.3 Relationship Between Classical and Proposed Models
Adopting the previous normalization, we are now in position to compare the time evolution and the equilibrium points of both models, in the particular, and still very important, time-invariant case by assuming that the pair of parameters \((\alpha (t), \gamma (t)) = (\alpha , \gamma )\) is constant for all \(t \in \mathbb {R}_+\). Dividing Eqs. (6) and (7) by (5), taking into account the basic reproduction number (9), we obtain the differential relations
First, set \(\nu =1\) corresponding to the classical SIR model index. Integrating (10) from \((s_0, i_0)\) to (s(t), i(t)) and (11) from \((s_0, r_0)\) to (s(t), r(t)) we obtain
while selecting the proposed SIR model with \(\nu =2\), these equations become
At the very beginning of the epidemic evolution, we have \(r_0=0\) and \(s_0 + i_0=1\) with \(s_0\) and \(i_0\) strictly positive initial conditions, such as the state variables remain positive for all \(t \in \mathbb {R}_+\). These initial conditions are called feasible. Moreover, from (5)–(7) it follows that any equilibrium point is such that \(i_*=0\) and \(s_* + r_* =1\) with \(s_*\) and \(r_*\) strictly positive.
The comparison of the time evolution of each model can be done by solving Eqs. (13) and (15), yielding
respectively. These solutions are virtually identical provided that \(s_0 \approx 1\) and \(0 < r(t) \ll 1\) which is always true in the beginning of the infectious process (even if \(R_0\) is large). However, as time passes, the solutions become different. It is important to mention that since both models include approximations, their validity must be established by comparison with data collected from reality. This aspect will be treated in a forthcoming section.
The previous equations, being true for all \(t \in \mathbb {R}_+\), characterize the trajectories of both models. They converge to equilibrium points, which always exist, provided that the initial condition is feasible. They can be calculated with no big difficulty. Indeed, performing the limit \(t \rightarrow +\infty \), Eqs. (12)–(13) give
that is, the determination of the equilibrium point follows from the solution of a transcendental equation. The same algebraic manipulations applied to Eqs. (14)–(15) provide
and it is seen that both admit strictly positive solutions \((s_*, r_*)\) whenever \(0< s_0 < 1\). Of course, depending on the value of the basic reproduction number, the number of susceptible at equilibrium \(s_*\) predicted by each model can be quite different. Interestingly, it can be verified that, at equilibrium, the number of susceptible (those not infected during the whole epidemic time evolution) predicted by the proposed SIR model is always bigger than the one provided by the classical SIR model. In other words, the time evolution of the epidemic process is always described as being less severe by the proposed SIR model. Once again, it is important to stress that this aspect needs factual confirmation.
4 The Unified Model in Discrete Time
For the COVID-19 outbreak, daily data is the only available data for parameter estimation. The Brazilian Ministry of Health and the World Health Organization daily report the number of new diagnosed cases \(n_m[k]\) and its accumulated sum \(a_m[k]\). Note that n[k] becomes very different from i[k] as the epidemic progresses. This is because infected people stop being infected when they transit to the removed type. The continuous-time independent variable \(t \in \mathbb {R}_+\) is evenly sampled providing the sampled instants of time \(t_k = k\Delta t\) where \(k \in \mathbb {N}\) is the discrete-time independent variable. As commented before, we have interest to consider \(\Delta t = 1~[\mathrm{day}]\).
The discrete-time version of the unified model is readily obtained by solving (approximately) the differential Eqs. (5)–(7) with the well-known Euler method, with unitary step size, which yields
It is interesting to notice that the equality \(s[k]+i[k]+r[k]=M\) holds for all \(k \in \mathbb {N}\) provided that it holds for \(k=0\), and the set of difference equations to be dealt with is nonlinear and time-varying. Furthermore, the additional equation
subject to the initial condition \(a[0]=a_m[0]\) generates the sequence of accumulated sum of new diagnosed cases predicted by the model, so as the minimization of the mean square error between the sequence a[k] and the measured sequence \(a_m[k]\) in some time interval provides the optimal time-varying parameters \(\alpha [k]\) and \(\gamma [k]\), for the time horizon under consideration.
Remark 2
Since \(\Delta t = 1~[\mathrm{day}]\), the discrete-time model admits two interpretations. First, \(\alpha [k]=\alpha (t_k)\) and \(\gamma [k]=\gamma (t_k)\) are expressed in \([\mathrm{day}^{-1}]\). Or alternatively, \(\alpha [k]=\alpha (t_k)\Delta t\) and \(\gamma [k]=\gamma (t_k)\Delta t\) are dimensionless.
Remark 3
It is simple to see that in discrete time, the unified model (20)–(22) follows without making use of Euler method arguments by adopting the same reasoning used to get the continuous-time counterpart. In this case, the parameter \(\gamma [k]=2 p[k] \in (0, 2)\) can also be interpreted as twice the probability that whenever a susceptible person meets an infected at \(k \in \mathbb {N}\), she/he becomes infected. In this sense, that parameter can also accommodate the possibility that several meetings between susceptible and infected occur in the time interval \([0, \Delta t]\). Notice that in this framework no kind of approximation is adopted in order to obtain the discrete-time version of the classical and proposed SIR models.
Remark 4
Assuming that in the discrete-time model there is no contagion then the epidemic must vanishes asymptotically which implies that \(\alpha [k] \in (0, 1)\).
Since the available measurement sequence \(a_m[k]\) belongs to the discrete-time domain, the mean square error does not take into account of the inter-sampling values \(a_m(t)\) for \(t \ne t_k, \forall k \in \mathbb {N}\). The consequence is that, provided the parameters are well determined, the final discrete-time model is, in general, a precise representation of the epidemic evolution but only at the sampling times \(t_k= k \Delta t,~\forall k \in \mathbb {N}\). In other words, as usual, the time evolution between the sampling instants \(\{t_k\}_{k \in \mathbb {N}}\) is not given by the discrete-time model.
4.1 Stability Analysis
Considering the normalization \(M=1\), which can be imposed with no loss of generality, let us define the closed and convex domain \(\mathbb {D}\subset \mathbb {R}^2\) that plays a central role in the stability analysis of SIR models, that is
and let us rewrite the previous model as \((s[k+1],i[k+1])= Q_\nu \circ (s[k],i[k])\) where the nonlinear operator \(Q_\nu : \mathbb {R}^2 \rightarrow \mathbb {R}^2\) given by
exhibits the important properties stated in the next lemma. Notice that the variable r[k] and Eq. (22) do not have importance, as far as stability is concerned.
Lemma 1
Assume that \(\alpha \in (0, 1)\). The set \(\mathbb {D}\subset \mathbb {R}^2\) is an invariant set of the nonlinear operator \(Q_\nu \), that is, \(Q_\nu \circ \mathbb {D}\subseteq \mathbb {D}\) provided that:
-
(i)
\(\nu = 2\) and \(\gamma \in (0, 3)\).
-
(ii)
\(\nu = 1\) and \(\gamma \in (0, 1)\).
Proof
Denoting \((s_Q,i_Q)= Q_\nu \circ (s,i)\), considering \(\alpha \in (0, 1)\) and \((s, i) \in \mathbb {D}\), it is immediate to verify that \(i_Q \ge 0\) and \(s_Q + i_Q = (s + i) - \alpha i \le 1\) for both \(\nu \in \{1, 2\}\). On the other hand, for any \(\gamma >0\), \(\nu \in \{1, 2\}\), and \((s, i) \in \mathbb {D}\) it is seen that \(s_Q = s - \gamma s^\nu i \ge s - \gamma s^\nu (1 - s)= g_\nu (s)\). Since \(g_\nu (0)=0\), two cases must be considered:
First, for \(\nu =2\), simple calculations put in evidence that \(g_\nu '(s) \ge 1- \gamma /3\). For \(\gamma \in (0, 3)\) the function \(g_\nu (s)\) is strictly increasing in the interval \(s \in [0, 1]\). As a consequence \(s_Q \ge 0\) for all \((s, i) \in \mathbb {D}\). Second, for \(\nu =1\), it can be verified that \(g_\nu '(s) \ge 0\) for all \(s \in [0, 1]\), provided that \(|\gamma | < 1\). Hence, in the interval \(\gamma \in (0, 1)\), we have that \(s_Q \ge 0\) for all \((s, i) \in \mathbb {D}\). The proof is concluded. \(\square \)
From Lemma 1, it is clear that for the proposed SIR model the trajectories \((s[k], i[k]) \in \mathbb {D}\) for all \(k \in \mathbb {N}\) provided that \((s_0, i_0) \in \mathbb {D}\), since its parameters are such that \(\alpha \in (0, 1)\) and \(\gamma \in (0, 2)\). For the classical SIR model, the situation is more restrictive since this property is assured whenever the parameters satisfy \(\alpha \in (0, 1)\) and \(\gamma \in (0, 1)\). If this last condition is violated, it may occur that \(s[k] <0\) for some \(k \in \mathbb {N}\), in which case, the meaning and validity of the model is completely lost.
Lemma 2
Assume that the conditions of Lemma 1 hold. For any initial condition \((s_0, i_0) \in \mathbb {D}\) the trajectory \((s[k], i[k])_{k \in \mathbb {N}} \in \mathbb {D}\) converges to an equilibrium point \((s_*, i_*)\) such that \(0 \le s_* \le s_0\) and \(i_*=0\).
Proof
Since any equilibrium point solves \((s_*, i_*) = Q_\nu \circ (s_*, i_*)\), it follows that \(i_*=0\). On the other hand, from Lemma 1, it has been established that \((s, i) \in \mathbb {D}\) provides \((s_Q, i_Q) \in \mathbb {D}\), which implies that the inequalities \(0 \le s_Q \le s \le 1\) hold as well. Consequently, the sequence \(s[k],~k \in \mathbb {N}\) converges to some \(0 \le s_* \le 1\) because it is bounded below and non increasing putting in evidence that \(s_* \le s_Q\). Now consider the linear function \(v(s,i) = (s-s_*) + i\) which is a valid Lyapunov function candidate for trajectories evolving in the region \((s, i) \in \mathbb {D}_* = \mathbb {D}\cap \{ s \ge s_* \}\) of the phase plane. Simple algebraic manipulations yield
for all \((s,i) \in \mathbb {D}_*\), concluding thus the proof. \(\square \)
For illustration we have drawn the phase plane for some parameters of the classical and proposed SIR models in discrete time. Figure 1 has been determined for \(\gamma = 1.75\) and \(\alpha = 0.50\). On the left side part the phase plane of the classical SIR model (\(\nu =1\)) is shown. It is clearly seen that, as expected, \(\mathbb {D}\) is not an invariant set since the condition of Lemma 1 is violated. In this case, the stability property of Lemma 2 is no longer valid. For comparison, from the right side part of the same figure it is clear that \(\mathbb {D}\) is an invariant set for the proposed SIR model (\(\nu =2\)).
Figure 2 has been determined for \(\gamma = 0.75\) and \(\alpha = 0.50\). For both models, \(\mathbb {D}\) is confirmed as an invariant set. For this particular choice of parameters, it is interesting to notice the similar behavior of both models but with the number of susceptible at equilibrium \(s_*\) being larger for the proposed model when compared to the one of the classical model. Under the same epidemic conditions, the proposed model appears to draw a less severe situation as the classical model does. This aspect needs factual confirmation.
4.2 Parameter Identification
In this section, we consider the parameter identification problem where the goal is to determine the time-varying parameters \((\alpha [k], \gamma [k])\) and the initial condition from the available data \(a_m[k]\). The number of days for which data are available is denoted by \(N_m\). Defining the state space variable \(z[k] = [s[k]~i[k]~a[k]]' \in \mathbb {R}^3\) with a[k] being the accumulated number of new diagnosed cases, the coupling variable w[k] and the output variable \(y[k]=a[k]\), the classical and the proposed SIR model state space minimal realization can be written as
where the indicated matrices are
and the nonlinear function \(f_\nu (\cdot ):\mathbb {R}^3 \rightarrow \mathbb {R}_+\) is defined as
Finally, it is important to keep in mind that the initial condition \(z_0 = [s_0~i_0~a_0]' \in \mathbb {R}^3\) must be non-negative and such that \(s_0 + i_0 \le M\). This is indicated simply by \(z_0 \in Z_0\). Additionally, the notation \((\alpha , \gamma ) \in \Pi _\nu \) denotes the constraints \(0 \le \alpha \le 1\) and \(0 \le \gamma \le \nu \) whenever the classical SIR model (\(\nu =1\)) or the proposed SIR model (\(\nu =2\)) is concerned. Under these constraints, Lemmas 1 and 2 indicate that \(\mathbb {D}\) is an invariant set and convergence towards the equilibrium point is assured. In this section, two complementary situations are analysed. First, the parameters are supposed to be time-invariant which naturally imposes that they are constant during the entire time horizon of interest. Afterward, the time-varying case with constant by parts parameters is treated.
4.3 Time-Invariant Parameter Optimization
A well-known procedure for parameter identification is adopted. It consists on the minimization of the mean square error between the model and data through the optimal solution of the nonlinear mathematical programming problem
where y[k] is the output provided by the model (27)–(29) with \((\alpha [k], \gamma [k])=(\alpha , \gamma )\) for all \(k \in [0, N_m)\). It is worth mentioning that this problem is highly non-convex, and by consequence, only a local optimum is expected to be reached by any numerical procedure. Hence, this is obviously true for the method applied here (Mathworks 2005). See Geromel et al. (2002) for robust filtering in a simpler context.
4.4 Time-Varying Parameter Optimization
It is assumed that the time interval \([0, N_m)\) is split in N sub-intervals \(\{T_j \}_{j=1}^N\), without overlapping, such that \((\alpha [k], \gamma [k]) = (\alpha _j, \gamma _j)\) for all \(k \in T_j,~j = 1, \ldots , N\). In other words, in each time sub-interval, the pair of parameters to be determined remains constant. We have to solve
where as before, y[k] is the output provided by the model (27)–(29). Numerically speaking, this problem is similar to (32). The only difference between them is the number of variables to handle. Moreover, since the constraints \((\alpha _j, \gamma _j) \in \Pi _\nu \) for \(j =1, \ldots , N\) are decoupled, at the optimal solution, the minimum cost naturally satisfies \(e_{tv} \le e_{ti}\) because any solution to (32) is feasible to (33). Depending on the epidemic horizon \(N_m\) and the number of time sub-intervals N, (33) can be classified as a large scale non-convex programming problem which, in general, is hard to solve mainly if the time horizon \(N_m\) under consideration is large.
4.5 Sequential Forward Optimization
In the time-varying parameter optimization context, whenever new measurements are treated during the epidemic time evolution, in principle, the time evolution of the parameters can be modified as well. To preserve optimal past values, a strategy inspired on receding horizon seems to be well adapted; see Bemporad et al. (2002) for details. In other words, the future of the outbreak evolution can not modify the values of parameters in the past and present. This imposes causality to the parameter identification procedure. It can be stated by considering again that the parameters \((\alpha [k], \gamma [k]) = (\alpha _j, \gamma _j)\) for all \(k \in T_j,~j = 1, \ldots , N\) are constant by parts. At an arbitrary time sub-interval \(T_j\), we need to solve
where y[k] is the output provided by the model (27)–(29) starting from the initial condition \(z_0 = [(M - a_m(0))~a_m(0)~a_m(0)]' \in Z_0\) at the first time sub-interval \(j=1\) and considering the state \(z[k+1]\) at the end of \(T_j\) as the initial condition for the subsequent time sub-interval \(T_{j+1}\). Proceeding in this way, it is possible to determine the time-varying parameters \((\alpha [k], \gamma [k])\) for all \(k \in [0, N_m)\). Finally, the minimum square error
between the sequence a[k] determined through the proposed time-varying model and the corresponding measured values \(a_m[k]\), actually observed, gives an idea of the model adherence to data. Compared to the previous strategy this one is simpler but suboptimal implying that \(e_{tv} \le e_{so}\). However, in general, it has been verified that \(e_{so} < e_{ti}\) whenever the time sub-intervals \(T_j,~j=1, \ldots , N\) are appropriately chosen. It is important to notice that the number of unknown variables of problem (34) is two for any \(j =1, \ldots , N\) which makes it easier to solve than the global optimization problem (33). It can be solved with no big difficulty even though the time horizon \(N_m\) of interest is large.
Finally, from the previous results, it is important to mention that, with precaution, since estimation errors may be expressive, the epidemic short-term evolution can be estimated by keeping the parameters constant, and equal to the values identified during the last time sub-interval \(T_N\), that is, \((\alpha [k],\gamma [k]) = (\alpha _N, \gamma _N)\) for all \(k \ge N_m\).
5 Simulation and Validation
In this section, the outbreak that is taking place in Brazil and the one that occurred in the UK are modeled. First, the outbreak in Brazil, which is until now in franc expansion, is considered. Afterward, the epidemic in the UK, which already reached the end, is handled through the same models of the SIR class, in order to put in evidence the adherence to data and precision. It is important to mention that in all cases the parameters have been determined by the sequential forward optimization method from the numerical solution of the nonlinear least square problem (34) with MATLAB Version R14, see Mathworks (2005). To avoid undesirable numerical singularity on the determination of the basic reproduction number \(R_0[k]=\gamma [k]/\alpha [k]\) we have replaced in the set \(\Pi _\nu \), the constraint \(0 \le \alpha \le 1\) by the constraint \(1/10 \le \alpha \le 1\). This is possible because for such a small enough lower bound the minimum mean square errors \(e_j^2,~j=1, \ldots , N\) remain approximately unchanged.
5.1 Outbreak in Brazil
In this study, we have adopted the official dataFootnote 4 available in the Brazilian Ministry of Health (2020) and World Health Organization (WHO) (2020) corresponding to the time horizon of 419 days since the first case in Brazil was reported. We have considered \(M \approx 210\) million inhabitants as indicated in Instituto Brasileiro de Geografia e Estatística (IBGE) (2020). The optimal parameters have been determined with \(N_m=419~[\mathrm{day}]\), \(N=28\) and \(T_1=[0, 28)\), \(T_j=[14j, 14(j+1)),~j=2, \ldots , 27\), \(T_{28}=[392, 419)\). Each time sub-interval was taken greater or equal to \(14~[\mathrm{day}]\) at exception to the first and the last ones. The larger length of the first sub-interval has been verified to be necessary to cope with the exponential growth of the epidemic at the very beginning. We have determined the optimal parameters corresponding to the classical SIR model (\(\nu = 1\)) and to the proposed SIR model (\(\nu =2\)). The residual error \(e_{so} \approx 4.24\) for both models indicates that they represent the data with similar accuracy. Accordingly, Fig. 3 shows the time evolution of the basic reproduction number \(R_0[k]\) in solid line (proposed model) and in dotted line (classical model) which are almost coincident in all days of the optimization horizon. The same framework is viewed in Fig. 4 which clearly shows that the accumulated sum of new diagnosed people a[k] provided by each model fit adequately and equally to data shown in red dots. This is not surprising since, until this moment, the number of susceptible individuals is close to the entire Brazilian population (\(\approx 93 \%\)), implying that \(R_0[k] \approx R_\nu [k]\) for all \(k \in [0, N_m)\) and \(\nu \in \{1, 2\}\), see (9).
Now we want to validate the two models. To this end, let us evaluate the sensitivity (robustness) of the optimal parameters against data errors. To this purpose, we have included one more day at the beginning of the outbreak with one infected person. Hence for \(N_m = 420\) we have set \(N=29\) and the same time sub-intervals \(T_j,~j=1, \ldots , 28\) and \(T_{29} = [406, 420)\). This means that the first 27 time sub-intervals are identical in both situations. We have determined the optimal parameters with the same algorithm and the same procedure. The following claims are supported by the trajectories shown in Figs. 5 and 6, respectively:
-
As before, in both figures we have plotted the trajectories provided by the proposed model (solid lines) and by the classical model (dotted lines). It is seen that they virtually coincide one to another and the same is true if we compare the accumulated sum of newly diagnosed people a[k] shown in the bottom of Fig. 4 and in the bottom of Fig. 6. This is in part explained by the fact that the residual errors are almost equal, namely \(e_{so} \approx 4.15\) and \(e_{so} \approx 4.17\), respectively. Hence, it appears that as far as the number of the accumulated sum of new infected is concerned, both models perform well and are numerically robust.
-
However, if we compare the basic reproduction number \(R_0[k]\) shown in Figs. 3 and 5 they appear completely different mainly at the beginning and at the middle of the outbreak time evolution. A possible explanation stems from the non-convex nature of the problem (34) which certainly has many local optima. Summarizing, it appears that the important index \(R_0[k]\) is not robust in face of the data perturbation considered. Hence, in the context of the proposed and the classical SIR models the estimation of the basic reproduction number must be done with care by paying particular attention to optimization methods and data quality.
As a final remark, we would like to put in evidence the fact that the outbreak in Brazil is composed by different and successive waves whose shape can not be described with precision by time-invariant models. Under unchanged conditions, both models estimate the end of the outbreak in Brazil by August, 2021.
5.2 Outbreak in the United Kingdom
We now move our attention to the outbreak in the United Kingdom that already reached the end after 441 days. Data for the entire time evolution of the epidemic is available in World Health Organization (WHO) (2020) and, consequently, all stages can be taken into account for parameter identification. We have considered a population of \(M \approx 68\) million inhabitants. As before, the optimal parameters have been determined with \(N_m=441~[\mathrm{day}]\), \(N=30\) and \(T_1=[0, 28)\), \(T_j=[14j, 14(j+1)),~j=2, \ldots , 29\), \(T_{30}=[420, 441)\). The residual error \(e_{so} \approx 3.68\) for both models indicates that they represent the data with similar accuracy. The total of accumulated new cases is very small (less than \(6.5 \%\)) when compared to the population, which naturally means that both models with \(\nu =1\) or \(\nu =2\) provide virtually the same trajectories.
Figure 7 shows the basic reproduction number for the proposed model in solid line and for the classical model in dotted line. Most of the time they are equal but in some instances they are quite different. Even though, as Fig. 8 makes clear, the trajectories for n[k] and a[k] are very similar, in fact, they are practically coincident. This indicates once again that the parameter estimation problem exhibits many local solutions which yield models of comparable precision. The non-convex nature of this problem needs supplementary research efforts towards the characterization of the global solution. This perhaps is possible if we take into account that the model nonlinearities are of polynomial type.
Finally, it is important to remark that the classical model and the proposed model of the SIR class are precise enough for analysis if we take care of their limitations to estimate the basic reproduction number. This aspect can also be improved by involving the determination of the time sub-intervals \(T_j,~j = 1, \ldots , N\) in the optimization process. Moreover, as clearly indicated in Costa et al. (2020) more research effort must also be done towards the development of more accurate prediction models depending on time-varying parameters. Another possibility, successfully adopted in Giordano et al. (2020), is to consider scenarios defined by parameters leading to prescribed \(R_0[k]\) and evaluating by the model the impact of each scenario on the epidemic evolution. To accomplish this goal, more precise and robust time-varying models are essential, see also Bertozzi et al. (2002).
6 Conclusion
The dynamic model proposed in this paper has good adherence to reality, however, due to the presence of time-varying parameters, it does not allow reliable long-term prediction of the epidemic time evolution. After all, if the parameters change in the future, there is no way to estimate them from observations of the past and present. Fortunately, the reported results seem to indicate that this fact becomes less important as new data is processed by the sequential optimization method which preserves causality and makes possible short-term prediction. Hence, the impact of well-defined scenarios on the epidemic evolution can be done with accuracy. For long-term prediction time-varying parameters modeling seems to be essential to increase precision. It is important to mention that we proposed an alternative dynamic model of the SIR class that, in principle, can be generalized to obtain continuous and discrete-time models like SEIR, SEIRS, SIRS, SEI, SEIS, SI, SIS, among others. Moreover, the robustness to cope with data errors, inevitable in practice, is a subject that needs further theoretical developments. Finally, we would like to emphasize once again that the practical viability of the proposed model needs factual confirmation.
Notes
It is interesting to put in evidence that this paper is until nowadays the most cited paper in the Proceedings of the Royal Society—A.
We would like to emphasize that we are not considering the possibility that people in the removed group may become susceptible again and be infected more often, as there is not enough evidence for this yet.
Since the population is big enough (\(M \gg 1\)), the pair of individuals can be chosen sequentially without replacement.
Data from both cited sources are slightly different. We have used those provided by the Brazilian Ministry of Health.
References
Anastassopoulou, C., Russo, L., Tsakris, A., & Siettos, C. (2020). Data-based analysis, modelling and forecasting of the COVID-19 outbreak. PLoS ONE,. https://doi.org/10.1371/journal.pone.0230405.
Anderson, R. M. (1987). The population dynamics of infectious diseases: Theory and applications. Springer.
Bailey, N. T. J. (1975). The mathematical theory of infectious diseases and its applications (2nd ed.), Hafner Press.
Bemporad, A., Borrelli, F., & Morari, M. (2002). Model predictive control based on linear programming: The explicit solution. IEEE Transactions on Automatic Control, 47, 1974–1985.
Bertozzi, A. L., Franco, E., Mohler, G., Short, M. B., & Sledge, D. (2002). The challenges of modeling and forecasting the spread of COVID-19. Proceedings of the National Academy of Sciences, 117, 16732–16738.
Brauer, F. & Castillo-Chavez, C. (2012). Mathematical models in population biology and epidemiology. Springer.
Brazilian Ministry of Health. (2020). Painel coronavirus.
Calafiore, G. C., Novara, C., & Possieri, C. (2020). A time-varying SIRD model for the COVID-19 contagion in Italy. Annual Reviews in Control, 50, 361–372.
Costa, J. A, Jr., Martinez, A. C., & Geromel, J. C. (2020). On an alternative susceptible-infected-removed epidemic model in discrete-time. Anais do Congresso Brasileiro de Automática,. https://doi.org/10.48011/asba.v2i1.995.
Geromel, J. C., de Oliveira, M. C., & Bernussou, J. (2002). Robust filtering of discrete-time linear systems with parameter dependent Lyapunov functions. SIAM Journal on Control and Optimization, 41, 700–711.
Giordano, G., Blanchini, F., Bruno, R., Colaneri, P., Di Filippo, A., Di Matteo, A., et al. (2020). Modelling the COVID-19 epidemic and implementation of population-wide interventions in Italy. Nature Medicine, 26, 855–860.
Hethcote, H. W. (2000). The mathematics of infectious diseases. SIAM Review, 42, 599–653.
Hu, Z., Teng, Z., & Zhang, L. (2014). Stability and bifurcation analysis in a discrete SIR epidemic model. Mathematics and Computers in Simulation, 97, 80–93.
Instituto Brasileiro de Geografia e Estatística (IBGE). (2020). Área territorial brasileira e população estimada.
Kermack, W. O., & McKendrick, A. G. (1927). A contribution to the mathematical theory of epidemics. Proceedings of the Royal Society of London. Series A, 115, 700–721.
Mathworks, Inc. (2005). MATLAB, Version R14 (7.1.0.246). Retrieved April 2020, from http://www.mathworks.com/products/matlab.html.
Silva, C. R. R., Almeida, A. C. L., Cardoso, R. T. N., & Takahashi, R. H. C. (2020). Epidemic individual-based models applied in random and scale-free networks. Revista Brasileira de Biometria, 38, 102–124.
Soares, C. D. (2011). Modelagem Matemática de Doenáas Infecciosas Considerando Heterogeneidade Etária: Estudo de Caso de Rubéola no México. Dissertação de Mestrado (in Portuguese), IMECC—UNICAMP.
World Health Organization (WHO). (2020). Coronavirus disease (COVID-2019) situation reports.
Acknowledgements
An early version of this paper was presented at XXIII Congresso Brasileiro de Automática (CBA 2020). The authors would like to thank the reviewers for their comments and concerns.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq/Brazil) under Grants 302013/2019-9 and 141289/2018-0.
Rights and permissions
About this article
Cite this article
Costa, J.A., Martinez, A.C. & Geromel, J.C. On the Continuous-time and Discrete-Time Versions of an Alternative Epidemic Model of the SIR Class. J Control Autom Electr Syst 33, 38–48 (2022). https://doi.org/10.1007/s40313-021-00757-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40313-021-00757-2