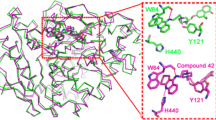
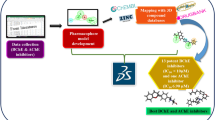
Abstract
Alzheimer’s disease (AD) is a leading cause of dementia in elderly patients. The pathophysiology of AD includes various pathways, such as the degradation of acetylcholine, amyloid-beta deposition, neurofibrillary tangle formation, and neuroinflammation. Many studies showed that targeting acetylcholinesterase enzyme (AChE) to improve acetylcholine can be an effective option to treat AD. In the current work, we employed a 3D QSAR-based approach to generate a pharmacophore to screen a chemical library of compounds that may inhibit AChE. Data from experimental studies were collected and used for the generation of pharmacophores. More than 1 million compounds were screened, and further drug-like properties were determined via in-silico ADMET studies. Techniques like molecular docking and molecular dynamics simulation were performed to analyze the binding of novel AChE inhibitors. A novel AChE inhibitor ligand-1 was identified as best with a docking score of -13.560 kcal/mol with RMSD of 1.71 Å during a 100 ns MD run. Further biological studies can give an insight into the potential of ligand-1 as a therapeutic agent for AD.
Graphical Abstract











Similar content being viewed by others
Availability of data and materials
Supplementary information (SI) is available for all relevant data free of charge via the Internet at https://doi.org/10.1007/s40203-024-00189-1.
Abbreviations
- AD:
-
Alzheimer’s disease
- ACh:
-
Acetylcholine
- AChE:
-
Acetylcholinesterase
- ADMET:
-
Absorption, distribution, metabolism, excretion and toxicity
- CAS:
-
Catalytic active site
- HBA:
-
Hydrogen bond acceptor
- HY:
-
Hydrophobic
- MD:
-
Molecular dynamics
- PAS:
-
Peripheral anionic site
- POS:
-
PosIonizable: Positive ionisable
- RA:
-
Ring aromatic
- RMSD:
-
Root mean square deviation
- RMSF:
-
Root mean square fluctuations
References
Akıncıoğlu H, Gülçin İ (2020) Potent acetylcholinesterase inhibitors: potential drugs for Alzheimer’s disease. Mini Rev Med Chem 20(8):703–715. https://doi.org/10.2174/1389557520666200103100521
Bai R, Guo J, Ye X-Y, Xie Y, Xie T (2022) Oxidative stress: the core pathogenesis and mechanism of Alzheimer’s disease. Ageing Res Rev 77:101619
BIOVIA, Dassault Systèmes, BIOVIA Discovery Studio 2022; San Diego.
Cheung GW, Cooper-Thomas HD, Lau RS, Wang LC (2023) Reporting reliability, convergent and discriminant validity with structural equation modeling: a review and best-practice recommendations. Asia Pac J Manag. https://doi.org/10.1007/s10490-023-09871-y
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7(1):42717. https://doi.org/10.1038/srep42717
Elgazar AA, El-Domany RA, Eldehna WM, Badria FA (2023) Theophylline-based hybrids as acetylcholinesterase inhibitors endowed with anti-inflammatory activity: synthesis, bioevaluation, in silico and preliminary kinetic studies. RSC Adv 13(36):25616–25634
Fernández-Bolaños JG, López Ó (2022) Butyrylcholinesterase inhibitors as potential anti-Alzheimer’s agents: an updated patent review (2018-present). Expert Opin Ther Pat 32(8):913–932
Ganji M, Bakhshi S, Shoari A, Ahangari Cohan R (2023) Discovery of potential FGFR3 inhibitors via QSAR, pharmacophore modeling, virtual screening and molecular docking studies against bladder cancer. J Transl Med 21(1):111
Gao H, Jiang Y, Zhan J, Sun Y (2021) Pharmacophore-based drug design of AChE and BChE dual inhibitors as potential anti-Alzheimer’s disease agents. Bioorg Chem 114(March):105149. https://doi.org/10.1016/j.bioorg.2021.105149
Jalali Z, Nejad Ebrahimi S, Rezadoost H (2023) Identifying natural products for gastric cancer treatment through pharmacophore creation, 3D QSAR, virtual screening, and molecular dynamics studies. DARU J Pharm Sci 31:243-258. https://doi.org/10.1007/s40199-023-00480-0
Jang C, Yadav DK, Subedi L, Venkatesan R, Venkanna A, Afzal S, Lee E, Yoo J, Ji E, Kim SY, Kim M (2018) Identification of novel acetylcholinesterase inhibitors designed by pharmacophore-based virtual screening, molecular docking and bioassay. Sci Rep 8(1):14921. https://doi.org/10.1038/s41598-018-33354-6
Jejurikar BL, Rohane SH (2021) Drug designing in discovery studio. Asian J Res Chem 14(2):135–138. https://doi.org/10.5958/0974-4150.2021.00025.0
Joy A, Menon S, Thomas NM, Christy M, Menon AD, John A (2023) Pharmacophore modelling and molecular dynamics simulation to identify novel molecules targeting catechol-O-methyltransferase and dopamine d3 receptor to combat Parkinson’s disease. Polym Bull. https://doi.org/10.1007/s00289-023-05087-8
Khan SS, Khatik LG, Datusalia KA (2023) Strategies for treatment of disease-associated dementia beyond Alzheimer’s disease: an update. Curr Neuropharmacol. https://doi.org/10.2174/1570159X20666220411083922
Kumar A, Rathi E, Kini SG (2020) Identification of potential tumour-associated carbonic anhydrase isozyme IX inhibitors: atom-based 3D-QSAR modelling, pharmacophore-based virtual screening and molecular docking studies. J Biomol Struct Dyn 38(7):2156–2170. https://doi.org/10.1080/07391102.2019.1626285
Liang J, Xu Z, Zhao Y (2022) Improved random batch Ewald method in molecular dynamics simulations. J Phys Chem A 126(22):3583–3593
LigPrep. Schrödinger Release 2014-2: LigPrep, Version 3.0. Schrödinger, LLC New York, NY 2014.
Liu Y, Zhang D, Tang Y, Gong X, Zheng J (2023) Development of a radical polymerization algorithm for molecular dynamics simulations of antifreezing hydrogels with double-network structures. NPJ Comput Mater 9(1):209
Lu C, Wu C, Ghoreishi D, Chen W, Wang L, Damm W, Ross GA, Dahlgren MK, Russell E, Von Bargen CD (2021) OPLS4: improving force field accuracy on challenging regimes of chemical space. J Chem Theory Comput 17(7):4291–4300
Nichols E, Steinmetz JD, Vollset SE, Fukutaki K, Chalek J, Abd-Allah F, Abdoli A, Abualhasan A, Abu-Gharbieh E, Akram TT, Al Hamad H, Alahdab F, Alanezi FM, Alipour V, Almustanyir S, Amu H, Ansari I, Arabloo J, Ashraf T, Astell-Burt T, Ayano G, Ayuso-Mateos JL, Baig AA, Barnett A, Barrow A, Baune BT, Béjot Y, Bezabhe WMM, Bezabih YM, Bhagavathula AS, Bhaskar S, Bhattacharyya K, Bijani A, Biswas A, Bolla SR, Boloor A, Brayne C, Brenner H, Burkart K, Burns RA, Cámera LA, Cao C, Carvalho F, Castro-de-Araujo LFS, Catalá-López F, Cerin E, Chavan PP, Cherbuin N, Chu D-T, Costa VM, Couto RAS, Dadras O, Dai X, Dandona L, Dandona R, De la Cruz-Góngora V, Dhamnetiya D, Dias da Silva D, Diaz D, Douiri A, Edvardsson D, Ekholuenetale M, El Sayed I, El-Jaafary SI, Eskandari K, Eskandarieh S, Esmaeilnejad S, Fares J, Faro A, Farooque U, Feigin VL, Feng X, Fereshtehnejad S-M, Fernandes E, Ferrara P, Filip I, Fillit H, Fischer F, Gaidhane S, Galluzzo L, Ghashghaee A, Ghith N, Gialluisi A, Gilani SA, Glavan I-R, Gnedovskaya EV, Golechha M, Gupta R, Gupta VB, Gupta VK, Haider MR, Hall BJ, Hamidi S, Hanif A, Hankey GJ, Haque S, Hartono RK, Hasaballah AI, Hasan MT, Hassan A, Hay SI, Hayat K, Hegazy MI, Heidari G, Heidari-Soureshjani R, Herteliu C, Househ M, Hussain R, Hwang B-F, Iacoviello L, Iavicoli I, Ilesanmi OS, Ilic IM, Ilic MD, Irvani SSN, Iso H, Iwagami M, Jabbarinejad R, Jacob L, Jain V, Jayapal SK, Jayawardena R, Jha RP, Jonas JB, Joseph N, Kalani R, Kandel A, Kandel H, Karch A, Kasa AS, Kassie GM, Keshavarz P, Khan MAB, Khatib MN, Khoja TAM, Khubchandani J, Kim MS, Kim YJ, Kisa A, Kisa S, Kivimäki M, Koroshetz WJ, Koyanagi A, Kumar GA, Kumar M, Lak HM, Leonardi M, Li B, Lim SS, Liu X, Liu Y, Logroscino G, Lorkowski S, Lucchetti G, Lutzky Saute R, Magnani FG, Malik AA, Massano J, Mehndiratta MM, Menezes RG, Meretoja A, Mohajer B, Mohamed Ibrahim N, Mohammad Y, Mohammed A, Mokdad AH, Mondello S, Moni MAA, Moniruzzaman M, Mossie TB, Nagel G, Naveed M, Nayak VC, Neupane Kandel S, Nguyen TH, Oancea B, Otstavnov N, Otstavnov SS, Owolabi MO, Panda-Jonas S, Pashazadeh Kan F, Pasovic M, Patel UK, Pathak M, Peres MFP, Perianayagam A, Peterson CB, Phillips MR, Pinheiro M, Piradov MA, Pond CD, Potashman MH, Pottoo FH, Prada SI, Radfar A, Raggi A, Rahim F, Rahman M, Ram P, Ranasinghe P, Rawaf DL, Rawaf S, Rezaei N, Rezapour A, Robinson SR, Romoli M, Roshandel G, Sahathevan R, Sahebkar A, Sahraian MA, Sathian B, Sattin D, Sawhney M, Saylan M, Schiavolin S, Seylani A, Sha F, Shaikh MA, Shaji KS, Shannawaz M, Shetty JK, Shigematsu M, Shin Il J, Shiri R, Silva DAS, Silva JP, Silva R, Singh JA, Skryabin VY, Skryabina AA, Smith AE, Soshnikov S, Spurlock EE, Stein DJ, Sun J, Tabarés-Seisdedos R, Thakur B, Timalsina B, Tovani-Palone MR, Tran BX, Tsegaye GW, Valadan Tahbaz S, Valdez PR, Venketasubramanian N, Vlassov V, Vu GT, Vu LG, Wang Y-P, Wimo A, Winkler AS, Yadav L, Yahyazadeh Jabbari SH, Yamagishi K, Yang L, Yano Y, Yonemoto N, Yu C, Yunusa I, Zadey S, Zastrozhin MS, Zastrozhina A, Zhang Z-J, Murray CJL, Vos T (2022) Estimation of the global prevalence of dementia in 2019 and forecasted prevalence in 2050: an analysis for the global burden of disease study 2019. Lancet Public Health. 7(2):e105–e125. https://doi.org/10.1016/S2468-2667(21)00249-8
Oviedo HF (2021) Implicit steepest descent algorithm for optimization with orthogonality constraints. Optim Lett 16:1773–1797
Pal S, Kumar V, Kundu B, Bhattacharya D, Preethy N, Reddy MP, Talukdar A (2019) Ligand-based pharmacophore modeling, virtual screening and molecular docking studies for discovery of potential topoisomerase I inhibitors. Comput Struct Biotechnol J 17:291–310. https://doi.org/10.1016/j.csbj.2019.02.006
Pirolli D, Righino B, Camponeschi C, Ria F, Di Sante G, De Rosa MC (2023) Virtual screening and molecular dynamics simulations provide insight into repurposing drugs against SARS-CoV-2 variants spike protein/ACE2 interface. Sci Rep 13(1):1494
Release S (2016) 4: Protein preparation wizard. Epik, Schrödinger, LLC, New York, NY, 2013–2018.
Reza-Zaldívar EE, Jacobo-Velázquez DA (2023) Comprehensive review of nutraceuticals against cognitive decline associated with Alzheimer’s disease. ACS Omega 8(39):35499–35522
Sarstedt M, Mooi E, Sarstedt M, Mooi E (2019) Regression Analysis. A concise guide to market research: the process, data, and methods using IBM SPSS Statistics. 209–256.
Sunseri J, Koes DR (2016) Pharmit: interactive exploration of chemical space. Nucleic Acids Res 44(W1):W442–W448
Tahami Monfared AA, Byrnes MJ, White LA, Zhang Q (2022) Alzheimer’s disease: epidemiology and clinical progression. Neurol Ther 11(2):553–569
Varma DA, Singh M, Wakode S, Dinesh NE, Vinaik S, Asthana S, Tiwari M (2023) Structure-based pharmacophore mapping and virtual screening of natural products to identify polypharmacological inhibitor against c-MET/EGFR/VEGFR-2. J Biomol Struct Dyn 41(7):2956–2970
Funding
None.
Author information
Authors and Affiliations
Contributions
HK carried out the research work by collecting the data and performing the computational studies. AKD interpreted the results, review of the manuscript. GLK hypothesized the concept and wrote and revised the manuscript. All the authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical Approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, H., Datusalia, A.K. & Khatik, G.L. Virtual screening of acetylcholinesterase inhibitors through pharmacophore-based 3D-QSAR modeling, ADMET, molecular docking, and MD simulation studies. In Silico Pharmacol. 12, 13 (2024). https://doi.org/10.1007/s40203-024-00189-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40203-024-00189-1