Abstract
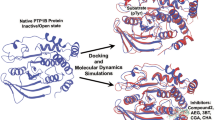
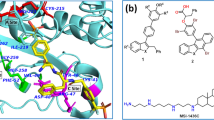
The present research scintillates on the homology modelling of rat mitochondrial protein tyrosine phosphatase 1 (PTPMT1) and targeting its activity using flavonoids through a computational docking approach. PTPMT1 is a dual-specificity phosphatase responsible for protein phosphorylation and plays a vital role in the metabolism of cardiolipin biosynthesis, insulin regulation, etc. The inhibition of PTPMT1 has also shown enhanced insulin levels. The three-dimensional structure of the protein is not yet known. The homology modelling was performed using SWISS-MODEL and Geno3D webservers to compare the efficiencies. The PROCHECK for protein modelled using SWISS-MODEL showed 91.6% of amino acids in the most favoured region, 0.7% residues in the disallowed region that was found to be significant compared to the model built using Geno3D. 210 common flavonoids were docked in the modelled protein using the AutoDock 4.2.6 along with a control drug alexidine dihydrochloride. Our results show promising candidates that bind protein tyrosine phosphatase 1, including, prunin (− 8.66 kcal/mol); oroxindin (− 8.56 kcal/mol); luteolin 7-rutinoside (− 8.47 kcal/mol); 3(2H)-isoflavenes (− 8.36 kcal/mol); nicotiflorin (− 8.29 kcal/mol), ranked top in the docking experiments. We predicted the pharmacokinetic and Lipinski properties of the top ten compounds with the lowest binding energies. To further validate the stability of the modelled protein and docked complexes molecular dynamics simulations were performed using Desmond, Schrodinger for 150 ns in conjunction with MM-GBSA. Thus, flavonoids could act as potential inhibitors of PTPMT1, and further, in-vitro and in-vivo studies are essential to complete the drug development process.
Graphical abstract


















Similar content being viewed by others
Availability of data and material
Not applicable.
Code availability
Not applicable.
Abbreviations
- PTPMT1:
-
Protein mitochondrial tyrosine phosphatase 1
- MM-GBSA:
-
Molecular mechanics-generalized born and surface area
- ATP:
-
Adenosine Tri Phosphate
- MD simulations:
-
Molecular dynamics simulations
- HIV:
-
Human Immunodeficiency Virus
- BLAST:
-
Basic local alignment search tool
- BLASTp:
-
Basic local alignment search tool protein
- FASTA:
-
FAST All
- GROMOS_96:
-
GROningen Molecular Simulation_96
- USA:
-
United States of America
- MGL Tools:
-
Molecular Graphic Laboratory tools
- PDB:
-
Protein data bank
- RMSD:
-
Root Mean Square Deviation
- RMSF:
-
Root Mean Square Fluctuation
- UCSF Chimera:
-
University of California San Francisco Chimera
- OPLS_2005:
-
Optimized Potentials for Liquid Simulations_2005
- TIP3P:
-
Transferable Intermolecular Potential with 3 Points
- GMQE:
-
Global Model Quality Estimation
- kcal/mol:
-
Kilocalories per mol
- IC50 :
-
Inhibitory Concentration 50
- µM:
-
Micromolar
- Å:
-
Angstroms
- Å2 :
-
Square Angstroms
- MTT:
-
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide
- μg GAE/mL:
-
Micrograms Gallic Acid Equivalent/millilitre
- mg/mL:
-
Milligrams per litter
- ns:
-
Nanoseconds
- mRNA:
-
Messenger Ribonucleic Acid
- Bcl-2:
-
B-cell lymphoma-2
- BAX:
-
Bcl-2-associated X protein
- ΔG:
-
Gibbs free energy
References
Abagyan R, Totrov M, Kuznetsov D (1994) ICM—a new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comput Chem 15:488–506. https://doi.org/10.1002/jcc.540150503
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Amin ML (2013) P-glycoprotein inhibition for optimal drug delivery. Drug Target Insights 7:27–34. https://doi.org/10.4137/DTI.S12519
Arthur DE, Ejeh S, Uzairu A (2020) Quantitative structure-activity relationship (QSAR) and design of novel ligands that demonstrate high potency and target selectivity as protein tyrosine phosphatase 1B (PTP 1B) inhibitors as an effective strategy used to model anti-diabetic agents. J Recept Signal Transd 40:501–520. https://doi.org/10.1080/10799893.2020.1759092
Azam SS, Abbasi SW (2013) Molecular docking studies for the identification of novel melatoninergic inhibitors for acetylserotonin-O-methyltransferase using different docking routines. Theor Biol Med Modell 10:63. https://doi.org/10.1186/1742-4682-10-63
Barthe L, Woodley J, Houin G (1999) Gastrointestinal absorption of drugs: methods and studies. Fundam Clin Pharmacol 13:154–168. https://doi.org/10.1111/j.1472-8206.1999.tb00334.x
Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350. https://doi.org/10.1093/bioinformatics/btq662
Bentivegna SS, Whitney KM (2002) Subchronic 3-month oral toxicity study of grape seed and grape skin extracts. Food Chem Toxicol 40:1731–1743. https://doi.org/10.1016/S0278-6915(02)00155-2
Brandman R, Brandman Y, Pande VS (2012) A-site residues move independently from P-site residues in all-atom molecular dynamics simulations of the 70S bacterial ribosome. PLoS One 7:e29377. https://doi.org/10.1371/journal.pone.0029377
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinf 10:421. https://doi.org/10.1186/1471-2105-10-421
Celiz G, Audisio MC, Daz M (2010) Antimicrobial properties of prunin, a citric flavanone glucoside, and its prunin 6″-O-lauroyl ester. J Appl Microbiol 109:1450–1457. https://doi.org/10.1111/j.1365-2672.2010.04773.x
Céliz G, Alfaro FF, Cappellini C, Daz M, Verstraeten SV (2013) Prunin-and hesperetin glucoside-alkyl (C4–C18) esters interaction with Jurkat cells plasma membrane: consequences on membrane physical properties and antioxidant capacity. Food Chem Toxicol 55:411–423. https://doi.org/10.1016/j.fct.2013.01.011
Chen P, Wu D, Pan Y (2013) Separation and purification of antioxidants from Ampelopsis heterophylla by counter-current chromatography. J Sep Sci 36:3660–3666. https://doi.org/10.1002/jssc.201300917
Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW, Tang Y (2019) admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J Chem Inf Model 59:4959. https://doi.org/10.1021/acs.jcim.9b00969
Cho N, Lee KY, Huh J, Choi JH, Yang H, Jeong EJ, Kim HP, Sung SH (2013) Cognitive-enhancing effects of Rhus verniciflua bark extract and its active flavonoids with neuroprotective and anti-inflammatory activities. Food Chem Toxicol 58:355–361. https://doi.org/10.1016/j.fct.2013.05.007
Choi JS, Yokozawa T, Oura H (1991) Antihyperlipidemic effect of flavonoids from Prunus davidiana. J Nat Prod 54:218–224. https://doi.org/10.1021/np50073a022
Cladis DP, Li S, Reddavari L, Cox A, Ferruzzi MG, Weaver CM (2020) A 90 day oral toxicity study of blueberry polyphenols in ovariectomized sprague-dawley rats. Food Chem Toxicol 139:111254. https://doi.org/10.1016/j.fct.2020.111254
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519. https://doi.org/10.1002/pro.5560020916
Combet C, Jambon M, Deleage G, Geourjon C (2002) Geno3D: automatic comparative molecular modelling of protein. Bioinformatics 18:213–214. https://doi.org/10.1093/bioinformatics/18.1.213
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717. https://doi.org/10.1038/srep42717
Day EK, Sosale NG, Lazzara MJ (2016) Cell signaling regulation by protein phosphorylation: a multivariate, heterogeneous, and context-dependent process. Curr Opin Biotechnol 40:185–192. https://doi.org/10.1016/j.copbio.2016.06.005
de Vasconcelos A, Campos VF, Nedel F, Seixas FK, Dellagostin OA, Smith KR, de Pereira CM, Stefanello FM, Collares T, Barschak AG (2013) Cytotoxic and apoptotic effects of chalcone derivatives of 2-acetyl thiophene on human colon adenocarcinoma cells. Cell Biochem Funct 31:289–297. https://doi.org/10.1002/cbf.2897
Doughty-Shenton D, Joseph JD, Zhang J, Pagliarini DJ, Kim Y, Lu D, Dixon JE, Casey PJ (2010) Pharmacological targeting of the mitochondrial phosphatase PTPMT1. J Pharmacol Exp Ther 333:584–592. https://doi.org/10.1124/jpet.109.163329
Filimonov DA, Druzhilovskiy DS, Lagunin AA, Gloriozova TA, Rudik AV, Dmitriev AV, Pogodin PV, Poroikov VV (2018) Computer-aided prediction of biological activity spectra for chemical compounds: opportunities and limitations. Biomed Chem 1:e00004. https://doi.org/10.18097/bmcrm00004
Fiser A, Sali A (2003) ModLoop: automated modeling of loops in protein structures. Bioinformatics 19:2500–2501. https://doi.org/10.1093/bioinformatics/btg362
Ganendren R, Widmer F, Singhal V, Wilson C, Sorrell T, Wright L (2004) In vitro antifungal activities of inhibitors of phospholipases from the fungal pathogen Cryptococcus neoformans. Antimicrob Agents Chemother 48:1561–1569. https://doi.org/10.1128/AAC.48.5.1561-1569.2004
Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A (2005) Protein identification and analysis tools on the ExPASy server. In: Walker JM (eds) The proteomics protocols handbook. Springer Protocols Handbooks. Humana Press. pp 571–607. https://doi.org/10.1385/1-59259-890-0:571
Genheden S, Ryde U (2015) The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov 10:449–461. https://doi.org/10.1517/17460441.2015.1032936
Gilbert P, Moore LE (2005) Cationic antiseptics: diversity of action under a common epithet. J Appl Microbiol 99:703–715. https://doi.org/10.1111/j.1365-2672.2005.02664.x
Goodarzi S, Tabatabaei MJ, Mohammad Jafari R, Shemirani F, Tavakoli S, Mofasseri M, Tofighi Z (2020) Cuminum cyminum fruits as source of luteolin-7-O-glucoside, potent cytotoxic flavonoid against breast cancer cell lines. Nat Prod Res 34:1602–1606. https://doi.org/10.1080/14786419.2018.1519824
Guan KL, Xiong Y (2011) Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci 36:108–116. https://doi.org/10.1016/j.tibs.2010.09.003
Guan K, Broyles SS, Dixon JE (1991) A Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature 350:359–362. https://doi.org/10.1038/350359a0
Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30:S162–S173. https://doi.org/10.1002/elps.200900140
Hafeez A, Saify ZS, Naz A, Yasmin F, Akhtar N (2013) Molecular docking study on the interaction of riboflavin (Vitamin) and cyanocobalamin (Vitamin) coenzymes. J Comput Med 2013:312183. https://doi.org/10.1155/2013/312183
Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL, Kaus JW (2016) OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12:281–296. https://doi.org/10.1021/acs.jctc.5b00864
Hari S, Akilashree S (2019) In silico homology modeling of presenilin 2-therapeutic target for Alzheimer’s disease. Res J Pharm Technol 12:3405–3409. https://doi.org/10.5958/0974-360X.2019.00575.4
He RJ, Yu ZH, Zhang RY, Zhang ZY (2014) Protein tyrosine phosphatases as potential therapeutic targets. Acta Pharmacol Sin 35:1227–1246. https://doi.org/10.1038/aps.2014.80
Hendlich M (1998) Databases for protein–ligand complexes. Acta Crystallogr Sect D 54:1178–1182. https://doi.org/10.1107/S0907444998007124
Hitchcock SA, Pennington LD (2006) Structure—brain exposure relationships. J Med Chem 49:7559–7583. https://doi.org/10.1021/jm060642i
Hollingsworth SA, Dror RO (2018) Molecular dynamics simulation for all. Neuron 99:1129–1143. https://doi.org/10.1016/j.neuron.2018.08.011
Horáková Ľ (2011) Flavonoids in prevention of diseases with respect to modulation of Ca-pump function. Interdiscip Toxicol 4:114–124. https://doi.org/10.2478/v10102-011-0019-5
Hospital A, Goñi JR, Orozco M, Gelpí JL (2015) Molecular dynamics simulations: advances and applications. Adv Appl Bioinf Chem 8:37–47. https://doi.org/10.2147/AABC.S70333
Huang B (2009) MetaPocket: a meta approach to improve protein ligand binding site prediction. OMICS 13:325–330. https://doi.org/10.1089/omi.2009.0045
Huang JL, Fu ST, Jiang YY, Cao YB, Guo ML, Wang Y, Xu Z (2007) Protective effects of Nicotiflorin on reducing memory dysfunction, energy metabolism failure and oxidative stress in multi-infarct dementia model rats. Pharmacol Biochem Behav 86:741–748. https://doi.org/10.1016/j.pbb.2007.03.003
Isah T (2019) Stress and defense responses in plant secondary metabolites production. Biol Res 52:39. https://doi.org/10.1186/s40659-019-0246-3
Jeffrey GA (1997) An introduction to hydrogen bonding. Oxford University Press, New York, p 228
Jendele L, Krivak R, Skoda P, Novotny M, Hoksza D (2019) PrankWeb: a web server for ligand binding site prediction and visualization. Nucleic Acids Res 47:W345–W349
Jung HA, Ali MY, Bhakta HK, Min BS, Choi JS (2017) Prunin is a highly potent flavonoid from Prunus davidiana stems that inhibits protein tyrosine phosphatase 1B and stimulates glucose uptake in insulin-resistant HepG2 cells. Arch Pharmacal Res 40:37–48. https://doi.org/10.1007/s12272-016-0852-3
Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II (2005) Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol 1:223–232. https://doi.org/10.1038/nchembio727
Kamaraj S, Ramakrishnan G, Anandakumar P, Jagan S, Devaki T (2009) Antioxidant and anticancer efficacy of hesperidin in benzo (a) pyrene induced lung carcinogenesis in mice. Invest New Drugs 27:214–222. https://doi.org/10.1007/s10637-008-9159-7
Kazuma K, Noda N, Suzuki M (2003) Malonylated flavonol glycosides from the petals of Clitoria ternatea. Phytochemistry 62:229–237. https://doi.org/10.1016/S0031-9422(02)00486-7
Kim JS, Kwon CS, Son KH (2000) Inhibition of alpha-glucosidase and amylase by luteolin, a flavonoid. Biosci Biotechnol Biochem 64:2458–2461. https://doi.org/10.1271/bbb.64.2458
Kraujalis P, Venskutonis PR, Kraujalienė V, Pukalskas A (2013) Antioxidant properties and preliminary evaluation of phytochemical composition of different anatomical parts of amaranth. Plant Foods Hum Nutr 68:322–328. https://doi.org/10.1007/s11130-013-0375-8
Kumar S, Pandey AK (2013) Chemistry and biological activities of flavonoids: an overview. Sci World J 2013:162750. https://doi.org/10.1155/2013/162750
Lai JP, Bao S, Davis IC, Knoell DL (2009) Inhibition of the phosphatase PTEN protects mice against oleic acid-induced acute lung injury. Br J Pharmacol 156:189–200. https://doi.org/10.1111/j.1476-5381.2008.00020.x
Lal Shyaula S, Abbas G, Siddiqui H, Sattar SA, Iqbal Choudhary M, Basha FZ (2012) Synthesis and antiglycation activity of kaempferol-3-O-rutinoside (nicotiflorin). Med Chem 8:415–420. https://doi.org/10.2174/1573406411208030415
Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486. https://doi.org/10.1007/BF00228148
Li M, Xu Z (2008) Quercetin in a lotus leaves extract may be responsible for antibacterial activity. Arch Pharmacal Res 31:640–644. https://doi.org/10.1007/s12272-001-1206-5
Li BW, Zhang FH, Serrao E, Chen H, Sanchez TW, Yang LM, Neamati N, Zheng YT, Wang H, Long YQ (2014) Design and discovery of flavonoid-based HIV-1 integrase inhibitors targeting both the active site and the interaction with LEDGF/p75. Bioorg Med Chem 22(12):3146–3158. https://doi.org/10.1016/j.bmc.2014.04.016
Lipinski CA (2000) Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods 44:235–249. https://doi.org/10.1016/S1056-8719(00)00107-6
Lipinski CA (2004) Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov Today 1:337–341. https://doi.org/10.1016/j.ddtec.2004.11.007
Liu Q, Zuo R, Wang K, Nong FF, Fu YJ, Huang SW, Pan ZF, Zhang Y, Luo X, Deng XL, Zhang XX (2020) Oroxindin inhibits macrophage NLRP3 inflammasome activation in DSS-induced ulcerative colitis in mice via suppressing TXNIP-dependent NF-κB pathway. Acta Pharmacol Sin 41:771–781. https://doi.org/10.1038/s41401-019-0335-4
Loureiro DR, Soares JX, Costa JC, Magalhães ÁF, Azevedo CM, Pinto MM, Afonso CM (2019) Structures, activities and drug-likeness of anti-infective xanthone derivatives isolated from the marine environment: a review. Molecules 24:243. https://doi.org/10.3390/molecules24020243
Massova I, Kollman PA (2000) Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspect Drug Discov Des 18:113–135. https://doi.org/10.1023/A:1008763014207
Morita O, Kirkpatrick JB, Tamaki Y, Chengelis CP, Beck MJ, Bruner RH (2009) Safety assessment of heat-sterilized green tea catechin preparation: a 6-month repeat-dose study in rats. Food Chem Toxicol 47:1760–1770. https://doi.org/10.1016/j.fct.2009.04.033
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Na EJ, Ryu JY (2018) Anti-inflammatory effects of prunin on UVB-irradiated human keratinocytes. Biomed Dermatol 2:14. https://doi.org/10.1186/s41702-018-0024-9
Nabae K, Hayashi SM, Kawabe M, Ichihara T, Hagiwara A, Tamano S, Tsushima Y, Uchida K, Koda T, Nakamura M, Ogawa K (2008) A 90-day oral toxicity study of purple corn color, a natural food colorant, in F344 rats. Food Chem Toxicol 46:774–780. https://doi.org/10.1016/j.fct.2007.10.004
Nahrstedt A, Hungeling M, Petereit F (2006) Flavonoids from Acalypha indica. Fitoterapia 77:484–486. https://doi.org/10.1016/j.fitote.2006.04.007
Ngo LT, Okogun JI, Folk WR (2013) 21st century natural product research and drug development and traditional medicines. Nat Prod Rep 30:584–592. https://doi.org/10.1039/C3NP20120A
Niemi NM, Lanning NJ, Westrate LM, MacKeigan JP (2013) Downregulation of the mitochondrial phosphatase PTPMT1 is sufficient to promote cancer cell death. PLoS One 8:e53803. https://doi.org/10.1371/journal.pone.0053803
Nishina A, Itagaki M, Suzuki Y, Koketsu M, Ninomiya M, Sato D, Suzuki T, Hayakawa S, Kuroda M, Kimura H (2017) Effects of flavonoids and triterpene analogues from leaves of Eleutherococcus sieboldianus (Makino) Koidz. ‘Himeukogi’in 3T3-L1 preadipocytes. Molecules 22:671. https://doi.org/10.3390/molecules22040671
Pagliarini DJ, Wiley SE, Kimple ME, Dixon JR, Kelly P, Worby CA, Casey PJ, Dixon JE (2005) Involvement of a mitochondrial phosphatase in the regulation of ATP production and insulin secretion in pancreatic β cells. Mol Cell 19:197–207. https://doi.org/10.1016/j.molcel.2005.06.008
Panche AN, Diwan AD, Chandra SR (2016) Flavonoids: an overview. J Nutr Sci 5:e47. https://doi.org/10.1017/jns.2016.41
Pant S, Singh M, Ravichandiran V, Murty US, Srivastava HK (2020) Peptide-like and small-molecule inhibitors against Covid-19. J Biomol Struct Dyn 39:1–10. https://doi.org/10.1080/07391102.2020.1757510
Park H, Lee J, Lee S (2006) Critical assessment of the automated AutoDock as a new docking tool for virtual screening. Proteins 65:549–554. https://doi.org/10.1002/prot.21183
Park H, Kim SY, Kyung A, Yoon TS, Ryu SE, Jeong DG (2012) Structure-based virtual screening approach to the discovery of novel PTPMT1 phosphatase inhibitors. Bioorg Med Chem Lett 22:1271–1275. https://doi.org/10.1016/j.bmcl.2011.10.083
Patterson KI, Brummer T, O’brien PM, Daly RJ (2009) Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J 418:475–489. https://doi.org/10.1042/BJ20082234
Pirvu L, Stefaniu A, Neagu G, Albu B, Pintilie L (2018) In vitro cytotoxic and antiproliferative activity of cydonia oblonga flower petals, leaf and fruit pellet ethanolic extracts. Docking simulation of the active flavonoids on anti-apoptotic protein Bcl-2. Open Chem 16:591–604. https://doi.org/10.1515/chem-2018-0062
Pitman MR, Ian Menz R (2006) Methods for protein homology modelling. Appl Mycol Biotechnol 6:37–59
Plazonić A, Bucar F, Maleš Ž, Mornar A, Nigović B, Kujundžić N (2009) Identification and quantification of flavonoids and phenolic acids in burr parsley (Caucalis platycarpos L.), using high-performance liquid chromatography with diode array detection and electrospray ionization mass spectrometry. Molecules 14:2466–2490. https://doi.org/10.3390/molecules14072466
Pourrat H, Bastide P, Dorier P, Pourrat A, Tranche P (1967) Preparation and therapeutic activity of some anthocyanin glycosides. Chim Ther 2:33–38
Pozzan A (2006) Molecular descriptors and methods for ligand based virtual high throughput screening in drug discovery. Cur Pharm Des 12:2099–2110. https://doi.org/10.2174/138161206777585247
Ramachandran GN, Ramakrishnan C, Sasisekharan V (1963) Stereochemistry of polypeptide chain configurations. J Mol Biol 7:95–99
Remmert M, Biegert A, Hauser A, Söding J (2012) HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9:173–175. https://doi.org/10.1038/nmeth.1818
Roccatano D, Barthel A, Zacharias M (2007) Structural flexibility of the nucleosome core particle at atomic resolution studied by molecular dynamics simulation. Biopolymers 85:407–421. https://doi.org/10.1002/bip.20690
Salvati AL, De Dominicis A, Tait S, Canitano A, Lahm A, Fiore L (2004) Mechanism of action at the molecular level of the antiviral drug 3 (2H)-isoflavene against type 2 poliovirus. Antimicrob Agents Chemother 48:2233–2243. https://doi.org/10.1128/AAC.48.6.2233-2243.2004
Schmid N, Christ CD, Christen M, Eichenberger AP, van Gunsteren WF (2012) Architecture, implementation and parallelisation of the GROMOS software for biomolecular simulation. Comput Phys Commun 183:890–903. https://doi.org/10.1016/j.cpc.2011.12.014
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385. https://doi.org/10.1093/nar/gkg520
Scott WR, Hünenberger PH, Tironi IG, Mark AE, Billeter SR, Fennen J, Torda AE, Huber T, Krüger P, van Gunsteren WF (1999) The GROMOS biomolecular simulation program package. J Phys Chem A 103:3596–3607. https://doi.org/10.1021/jp984217f
Shi Q, Li L, Huo C, Zhang M, Wang Y (2010) Study on natural medicinal chemistry and new drug development. Zhongcaoyao = Chinese Traditional and Herbal Drugs 41:1583–1589
Shivanika C, Kumar D, Ragunathan V, Tiwari P, Sumitha A (2020) Molecular docking, validation, dynamics simulations, and pharmacokinetic prediction of natural compounds against the SARS-CoV-2 main-protease. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1815584
Shoji T, Akazome Y, Kanda T, Ikeda M (2004) The toxicology and safety of apple polyphenol extract. Food Chem Toxicol 42:959–967. https://doi.org/10.1016/j.fct.2004.02.008
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Sinha S, Batovska DI, Medhi B, Radotra BD, Bhalla A, Markova N, Sehgal R (2019) In vitro anti-malarial efficacy of chalcones: cytotoxicity profile, mechanism of action and their effect on erythrocytes. Malar J 18:421. https://doi.org/10.1186/s12936-019-3060-z
Sirikantaramas S, Yamazaki M, Saito K (2008) Mechanisms of resistance to self-produced toxic secondary metabolites in plants. Phytochem Rev 7:467. https://doi.org/10.1007/s11101-007-9080-2
Steck PA, Pershouse MA, Jasser SA, Yung WA, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C (1997) Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23. 3 that is mutated in multiple advanced cancers. Nat Genet 15:356–362
Stewart JJ (1989) Optimization of parameters for semiempirical methods II. Applications. J Comput Chem 10:221–264. https://doi.org/10.1002/jcc.540100209
Torres MC, Pinto FD, Braz-Filho R, Silveira ER, Pessoa OD, Jorge RJ, Ximenes RM, Monteiro HS, Evangelista JS, Diz-Filho EB, Toyama MH (2011) Antiophidic solanidane steroidal alkaloids from Solanum campaniforme. J Nat Prod 74:2168–2173. https://doi.org/10.1021/np200479a
Vyas VK, Ukawala RD, Ghate M, Chintha C (2012) Homology modeling a fast tool for drug discovery: current perspectives. Indian J Pharm Sci 74:1–17. https://doi.org/10.4103/0250-474X.102537
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng Des Sel 8:127–134. https://doi.org/10.1093/protein/8.2.127
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TA, Rempfer C, Bordoli L, Lepore R (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. https://doi.org/10.1093/nar/gky427
Wu D, Kong Y, Han C, Chen J, Hu L, Jiang H, Shen X (2008) D-Alanine: D-alanine ligase as a new target for the flavonoids quercetin and apigenin. Int J Antimicrob Agents 32:421–426. https://doi.org/10.1016/j.ijantimicag.2008.06.010
Xiao J, Engel JL, Zhang J, Chen MJ, Manning G, Dixon JE (2011) Structural and functional analysis of PTPMT1, a phosphatase required for cardiolipin synthesis. Proc Natl Acad Sci USA 108:11860–11865. https://doi.org/10.1073/pnas.1109290108
Yadav R, Selvaraj C, Aarthy M, Kumar P, Kumar A, Singh SK, Giri R (2020) Investigating into the molecular interactions of flavonoids targeting NS2B-NS3 protease from ZIKA virus through in-silico approaches. J Biomol Struct Dyn 39:1–13. https://doi.org/10.1080/07391102.2019.1709546
Yuan H, Ma Q, Ye L, Piao G (2016) The traditional medicine and modern medicine from natural products. Molecules 21:559. https://doi.org/10.3390/molecules21050559
Zang Y, Igarashi K, Li Y (2016) Anti-diabetic effects of luteolin and luteolin-7-O-glucoside on KK-A y mice. Biosci Biotechnol Biochem 80:1580–1586. https://doi.org/10.1080/09168451.2015.1116928
Zhang L, Liu W, Hu T, Du L, Luo C, Chen K, Shen X, Jiang H (2008) Structural basis for catalytic and inhibitory mechanisms of β-hydroxyacyl-acyl carrier protein dehydratase (FabZ). J Biol Chem 283:5370–5379. https://doi.org/10.1074/jbc.M705566200
Zhang J, Guan Z, Murphy AN, Wiley SE, Perkins GA, Worby CA, Engel JL, Heacock P, Nguyen OK, Wang JH, Raetz CR (2011a) Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metab 13:690–700. https://doi.org/10.1016/j.cmet.2011.04.007
Zhang Z, Li Y, Lin B, Schroeder M, Huang B (2011b) Identification of cavities on protein surface using multiple computational approaches for drug binding site prediction. Bioinformatics 27:2083–2088. https://doi.org/10.1093/bioinformatics/btr331
Zhao BT, Nguyen DH, Le DD, Choi JS, Min BS, Woo MH (2018) Protein tyrosine phosphatase 1B inhibitors from natural sources. Arch Pharmacal Res 41:130–161. https://doi.org/10.1007/s12272-017-0997-8
Zheng X, Polli J (2010) Identification of inhibitor concentrations to efficiently screen and measure inhibition Ki values against solute carrier transporters. Eur J Pharm Sci 41:43–52. https://doi.org/10.1016/j.ejps.2010.05.013
Acknowledgements
The authors sincerely thank Alagappa College of Technology, Anna University, Chennai for towards the successful completion of research work. The authors also thank the National Institute of Pharmaceutical Education and Research, Kolkata for performing Molecular Dynamics simulations.
Funding
The authors did not receive any funding for the research from any funding bodies.
Author information
Authors and Affiliations
Contributions
VR: designed and performed the experiments and wrote the paper; KC: conceptualization, review, editing the manuscript; CS: performed the experiments and drafting; MSS: correcting and editing the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ragunathan, V., Chithra, K., Shivanika, C. et al. Modelling and targeting mitochondrial protein tyrosine phosphatase 1: a computational approach. In Silico Pharmacol. 10, 3 (2022). https://doi.org/10.1007/s40203-022-00119-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40203-022-00119-z