Abstract
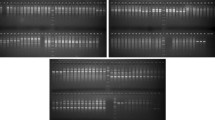
Jasmines are an important group of plants extensively used in the perfumery industry, preparation of garland and for ornamental purposes. To analyze the genetic potential of the group, the authors evaluated its genetic diversity. Intersimple sequence repeats (ISSR) markers were used to evaluate genetic diversity among 40 accessions of 23 Jasminum species including six endemic species. The present study is the first report of the efficacy of 10 shortlisted ISSR markers out of 30 primers screened. Among 23 Jasminum species, J. bignoniaceum revealed maximum genetic diversity and Shannon’s Index. Results revealed that the accessions of Jasminum species are grouped into three major clades, showing close agreement with the morphological relatedness. The diversity analysis also showed that the members of section Alternifolia are the ancestor to the rest of the Jasminum spp. The study provides direction for the future population study on Jasminum spp. This is the first report on the genetic diversity assessment of Indian Jasminum spp. incorporating approximately 50% species.






Similar content being viewed by others
References
Green PS (1969) Studies in the genus Jasminum L. IV. These so-called new world species. Kew Bull 23:273–275
Green PS, Miller D (2009) The genus Jasminum in cultivation. Kew Publishing, Royal Botanic Gardens, Kew
Green PS (2004) Oleaceae. In: Kadereit JW (ed) Flowering plants, dicotyledons: lamiales (except Acanthaceae including Avicenniaceae), vol 7. Springer, New York, pp 296–306
De Candolle AP (1844) Prodromus Systematis Naturalis Regni Vegetabilis 8. Treuttel and Wfirtz, Paris
Green PS (2001) Studies in the genus Jasminum, XVII: sections Trifoliolata and Primulina. Kew Bull 56:903–915
Jeyrani N, Yohanan R, Priya D, Dwivedi MD, Pandey AK (2018) Molecular systematics of Jasminum L. (Oleaceae) in India with discussion on evolution of leaf morphology. J Gen 97:1225–1239
Mukundan S (2000) Characterization of important cultivars of Jasminum species using molecular markers. M.Sc. thesis submitted to College of Agriculture, UAS, Bangalore-65
Besnard G, Khadari B, Villemur P, Bervill A (2000) Cytoplasmic male sterility in the olive (Olea europaea L.). Theor Appl Genet 100:1018–1024
Rosa R, Angiolillo A, Guerrero C, Pellegrini M, Rallo L, Besnard G, Bervill A, Martin A (2003) A first linkage map of olive (Olea europaea L.) cultivars using RAPD, AFLP, RFLP and SSR markers. Theor Appl Genet 106:1273–1282
Carriero F, Fontanazza G, Cellini F, Giorio G (2002) Identification of simple sequence repeats (SSRs) in olive (Olea europaea). Theor Appl Genet 104(2–3):301–307
Sensi E, Vignani R, Scali M, Masi E, Cresti M (2003) DNA fingerprinting and genetic relatedness among cultivated of Olea europaea L. estimated by AFLP analysis. Sci Hort 97:379–388
Lopes MS, Mendonca D, Sefc KM, Gil FS, Machado AC (2004) Genetic evidence of intra-cultivar variability within Iberian olive cultivars. Hort Sci 39:1562–1565
Hagidikitriou M, Katsiotis A, Menexes G, Pontikis C, Loukas M (2005) Genetic diversity of major Greek olive cultivars using molecular (AFLPs and RAPADs) markers and morphological traits. Am Soc Hort Sci 130:211–217
Mantia ML, Guerin J, Sedgley M, Barone E (2006) Identification of olive (Olea europaea L.) genotypes using SSR and RAPD markers. Biotechnologies et qualite des produits de I’olivier dans le bassin mediterraneen 10:9–14
Mahmood MA, Hafiz IA, Abbasi NA, Faheem M (2013) Detection of genetic diversity in Jasminum species through RAPD techniques. Int J Agric Biol 15:505–510
Shekhar S, Sriram S, Prasad MP (2013) Genetic diversity determination of jasmine species by DNA fingerprinting using molecular markers. Int J Biotechnol Bioeng Res 4:335–340
Ghehsareh G, Salehi MH, Khosh-Khui M et al (2015) Application of ISSR markers to analyze molecular relationships in Iranian Jasmine (Jasminum spp.) accessions. Mol Biotechnol 57:65. https://doi.org/10.1007/s12033-014-9802-9
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Paithankar KR, Prasad KSN (1991) Precipitation of DNA by polyethylene glycol and ethanol. Nucl Acids Res 19:1346. https://doi.org/10.1093/nar/19.6.1346
Yeh FC, Yang R-C, Boyle Timothy BJ, Ye Z-H, Mao Judy X (1997) POPGENE, the user-friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Canada
Perrier X, Jacquemound-Collet JP (2006) Darwin software. http://darwin.cirad.fr/darwin
Yan X, Xiao B, Han H, Yuan W, Shang F (2008) AFLP analysis of genetic relationships and diversity of some Chinese Osmanthus fragrans cultivars. Life Sci J 6:2
Amirmoradi B, Talebi R, Karami E (2012) Comparison of genetic variation and differentiation among annual Cicer species using start codon targeted (SCoT) polymorphism, DAMD-PCR, and ISSR markers. Plant Syst Evol 298(9):1679–1689. https://doi.org/10.1007/s00606-012-0669-6
McCoy TJ, Echt ES (1993) Potential of trispecies bridge crosses and random amplified polymorphic DNA markers for introgression of Medicago daghestanica and M. pironaegermplasm into alfalfa (M. sativa). Genome 36:594–601
Brown SM, Resovich SK (1996) In: Paterson AH (ed) Genome mapping in plants. Clandes, New York, pp 85–93
Joshi SP, Gupta VS, Aggarwal RK, Ranjekar PK, Brar DS (2000) Genetic diversity and phylogenetic relationship as revealed by inter simple sequence repeat (ISSR) polymorphism in the genus Oryza. Theor Appl Genet 100:1311–1320
Acknowledgements
The authors are thankful to UGC, New Delhi, for financial support (#32-409/2006 (SR), and to Kerala Forest Department for permission to collect Jasminum spp. from forest areas in Kerala. NJ, RY, DV greatly acknowledge the help rendered by the Principals of respective Sree Narayana Colleges at Chempazhanthy and Kollam for providing research facilities.
Author information
Authors and Affiliations
Contributions
AKP conceived the idea and provided all the laboratory facilities at the Department of Botany, University of Delhi and also helped in designing the experiments and finalizing the manuscript. RY collected specimens for herbarium preparation and preserved leaf materials for DNA extraction and also helped in experimental work and manuscript preparation. NJJ helped in collection of materials and manuscript writing. DV helped in experimental design, manuscript preparation, and final discussion. SAR helped in experimental work. SK helped in analysis of data and manuscript writing. JT helped in experimental work, data analysis, and manuscript writing. MDD helped in experimental design, laboratory work, interpretation of data, and manuscript writing.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest among the authors to publish this manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance Statement
Genus Jasminum is an economically important oil-yielding taxa. Despite its high economic value, no molecular study was done to assess the genetic diversity within the genus in India. ISSR-PCR technique will help to estimate the genetic similarities and evaluate relationships between available accessions of Jasminum species in India.
Rights and permissions
About this article
Cite this article
Yohanan, R., Jeyarani, N.J., Devipriya, V. et al. Evaluating Genetic Diversity Within Genus Jasminum L. (Oleaceae) Using Intersimple Sequence Repeats (ISSR) Marker. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 90, 531–540 (2020). https://doi.org/10.1007/s40011-019-01124-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-019-01124-7