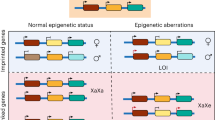
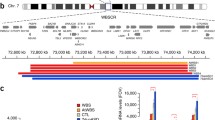
Abstract
Recent advances in stem cell biology have dramatically increased the understanding of molecular and cellular mechanism of pluripotency and cell fate determination. Additionally, pluripotent stem cells (PSCs), including embryonic stem cells and induced pluripotent stem cells, arose as essential resources for disease modeling and cellular therapeutics. Despite these advancements, the epigenetic dysregulation in pluripotency such as the imprinting status, and X chromosome dosage compensation, and its consequences on future utility of PSCs yet remain unresolved. In this review, we will focus on the X chromosome regulation in human PSCs (hPSCs). We will introduce the previous findings in the dosage compensation process on mouse model, and make comparison with those of human systems. Particularly, the X chromosome activation status of human preimplantation embryos, and the regulation of the active X chromosome by human specific lincRNA, X Active Coating Transcript (XACT), will be discussed. We will also discuss the recent findings on higher order X chromosome architecture, and abnormal X chromosome status in hPSCs.



Similar content being viewed by others
Change history
19 January 2018
Unfortunately, Acknowledgements section was missing in the originally published article.
References
Clarke D, Frisen J. Differentiation potential of adult stem cells. Curr Opin Genet Dev. 2001;11:575–80.
Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–53.
Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322(5903):945–9.
Jeong J, Kim KN, Chung MS, Kim HJ. Functional comparison of human embryonic stem cells and induced pluripotent stem cells as sources of hepatocyte-like cells. Tissue Eng Regen Med. 2016;13:740–9.
Hong K. Cellular reprogramming and its application in regenerative medicine. Tissue Eng Regen Med. 2015;12:80–9.
Okamoto I, Patrat C, Thépot D, Peynot N, Fauque P, Daniel N, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472:370–4.
Vallot C, Patrat C, Collier AJ, Huret C, Casanova M, Liyakat Ali TM, et al. XACT noncoding RNA competes with XIST in the control of X chromosome activity during human early development. Cell Stem Cell. 2017;20:102–11.
Petropoulos S, Edsgärd D, Reinius B, Deng Q, Panula SP, Codeluppi S, et al. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. 2016;167:285.
Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11(2):156–66.
Sangrithi MN, Royo H, Mahadevaiah SK, Ojarikre O, Bhaw L, Sesay A, et al. Non-canonical and sexually dimorphic X dosage compensation states in the mouse and human germline. Dev Cell. 2017;40:289–301.
Burgoyne PS, Biggers JD. The consequences of X-dosage deficiency in the germ line: impaired development in vitro of preimplantation embryos from XO mice. Dev Biol. 1976;51:109–17.
Theunissen TW, Friedli M, He Y, Planet E, O’Neil RC, Markoulaki S, et al. Molecular criteria for defining the naive human pluripotent state. Cell Stem Cell. 2016;19:502–15.
Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell. 2014;15:524–6.
Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153:1537–51.
Donohoe ME, Silva SS, Pinter SF, Xu N, Lee JT. The pluripotency factor Oct4 interacts with Ctcf and also controls X-chromosome pairing and counting. Nature. 2009;460:128–32.
Masui O, Bonnet I, Le Baccon P, Brito I, Pollex T, Murphy N, et al. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell. 2011;145:447–58.
Furlan G, Rougeulle C. Function and evolution of the long noncoding RNA circuitry orchestrating X-chromosome inactivation in mammals. Wiley Interdiscip Rev RNA. 2016;7:702–22.
Keniry A, Gearing LJ, Jansz N, Liu J, Holik AZ, Hickey PF, et al. Setdb1-mediated H3K9 methylation is enriched on the inactive X and plays a role in its epigenetic silencing. Epigenet Chromatin. 2016;9:16.
Minkovsky A, Patel S, Plath K. Concise review: pluripotency and the transcriptional inactivation of the female mammalian X chromosome. Stem Cells. 2012;30:48–54.
Chen CK, Blanco M, Jackson C, Aznauryan E, Ollikainen N, Surka C, et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354:468–72.
Vallot C, Ouimette JF, Rougeulle C. Establishment of X chromosome inactivation and epigenomic features of the inactive X depend on cellular contexts. Bioessays. 2016;38:869–80.
Ogawa Y, Lee JT. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell. 2003;11:731–43.
Makhlouf M, Ouimette JF, Oldfield A, Navarro P, Neuillet D, Rougeulle C. A prominent and conserved role for YY1 in Xist transcriptional activation. Nat Commun. 2014;5:4878.
Hanna J, Cheng AW, Saha K, Kim J, Lengner CJ, Soldner F, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A. 2010;107:9222–7.
Ware CB. Naive embryonic stem cells: the future of stem cell research? Regen Med. 2014;9:401–3.
Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–6.
Chan YS, Göke J, Ng JH, Lu X, Gonzales KA, Tan CP, et al. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell. 2013;13:663–75.
Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, et al. Derivation of naive human embryonic stem cells. Proc Natl Acad Sci U S A. 2014;111:4484–9.
Guo G, von Meyenn F, Santos F, Chen Y, Reik W, Bertone P, et al. Naive pluripotent stem cells derived directly from isolated cells of the human inner cell mass. Stem Cell Reports. 2016;6:437–46.
Lanphier E, Urnov F, Haecker SE, Werner M, Smolenski J. Don’t edit the human germ line. Nature. 2015;519:410–1.
Savulescu J, Pugh J, Douglas T, Gyngell C. The moral imperative to continue gene editing research on human embryos. Protein Cell. 2015;6:476–9.
Elstner A, Damaschun A, Kurtz A, Stacey G, Arán B, Veiga A, et al. The changing landscape of European and international regulation on embryonic stem cell research. Stem Cell Res. 2009;2:101–7.
Nielen MG, de Vries SA, Geijsen N. European stem cell research in legal shackles. EMBO J. 2013;32:3107–11.
Salter B, Salter C. Bioethical ambition, political opportunity and the European governance of patenting: the case of human embryonic stem cell science. Soc Sci Med. 2013;98:286–92.
Zarzeczny A, Caulfield T. Emerging ethical, legal and social issues associated with stem cell research and the current role of the moral status of the embryo. Stem Cell Rev. 2009;5:96–101.
Pera MF. Human embryo research and the 14-day rule. Development. 2017;144:1923–5.
Zhai X, Ng V, Lie R. No ethical divide between China and the West in human embryo research. Dev World Bioeth. 2016;16:116–20.
Pennings G, Segers S, Debrock S, Heindryckx B, Kontozova-Deutsch V, Punjabi U, et al. Human embryo research in Belgium: an overview. Fertil Steril. 2017;108:96–107.
Vallot C, Huret C, Lesecque Y, Resch A, Oudrhiri N, Bennaceur-Griscelli A, et al. XACT, a long noncoding transcript coating the active X chromosome in human pluripotent cells. Nat Genet. 2013;45:239–41.
Vallot C, Ouimette JF, Makhlouf M, Féraud O, Pontis J, Côme J, et al. Erosion of X chromosome inactivation in human pluripotent cells initiates with XACT coating and depends on a specific heterochromatin landscape. Cell Stem Cell. 2015;16:533–46.
Patel S, Bonora G, Sahakyan A, Kim R, Chronis C, Langerman J, et al. Human embryonic stem cells do not change their X inactivation status during differentiation. Cell Rep. 2017;18:54–67.
McDonald G, Cabal N, Vannier A, Umiker B, Yin RH, Orjalo AV Jr, et al. Female bias in systemic lupus erythematosus is associated with the differential expression of X-linked toll-like receptor 8. Front Immunol. 2015;6:457.
Sahakyan A, Kim R, Chronis C, Sabri S, Bonora G, Theunissen TW, et al. Human naive pluripotent stem cells model X chromosome dampening and X inactivation. Cell Stem Cell. 2017;20:87–101.
Gerdes P, Richardson SR, Mager DL, Faulkner GJ. Transposable elements in the mammalian embryo: pioneers surviving through stealth and service. Genome Biol. 2016;17:100.
Gifford WD, Pfaff SL, Macfarlan TS. Transposable elements as genetic regulatory substrates in early development. Trends Cell Biol. 2013;23:218–26.
Garcia-Perez JL, Widmann TJ, Adams IR. The impact of transposable elements on mammalian development. Development. 2016;143:4101–14.
Thompson PJ, Macfarlan TS, Lorincz MC. Long terminal repeats: from parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol Cell. 2016;62:766–76.
Chadwick BP. DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res. 2008;18:1259–69.
Giacalone J, Friedes J, Francke U. A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nat Genet. 1992;1:137–43.
Horakova AH, Calabrese JM, McLaughlin CR, Tremblay DC, Magnuson T, Chadwick BP. The mouse DXZ4 homolog retains Ctcf binding and proximity to Pls3 despite substantial organizational differences compared to the primate macrosatellite. Genome Biol. 2012;13:R70.
Deng X, Ma W, Ramani V, Hill A, Yang F, Ay F, et al. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 2015;16:152.
Darrow EM, Huntley MH, Dudchenko O, Stamenova EK, Durand NC, Sun Z, et al. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci U S A. 2016;113:E4504–12.
Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, et al. Structural organization of the inactive X chromosome in the mouse. Nature. 2016;535:575–9.
Yang F, Deng X, Ma W, Berletch JB, Rabaia N, Wei G, et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015;16:52.
Disteche CM. Escapees on the X chromosome. Proc Natl Acad Sci U S A. 1999;96:14180–2.
Slavney A, Arbiza L, Clark AG, Keinan A. Strong constraint on human genes escaping X-inactivation is modulated by their expression level and breadth in both sexes. Mol Biol Evol. 2016;33:384–93.
Chaligné R, Heard E. X-chromosome inactivation in development and cancer. FEBS Lett. 2014;588:2514–22.
Chaligné R, Popova T, Mendoza-Parra MA, Saleem MA, Gentien D, Ban K, et al. The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res. 2015;25:488–503.
Peeters SB, Cotton AM, Brown CJ. Variable escape from X-chromosome inactivation: identifying factors that tip the scales towards expression. Bioessays. 2014;36:746–56.
Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, et al. Molecular maps of the reorganization of genome–nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–13.
Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5.
Collier AJ, Panula SP, Schell JP, Chovanec P, Plaza Reyes A, Petropoulos S, et al. Comprehensive cell surface protein profiling identifies specific markers of human naive and primed pluripotent states. Cell Stem Cell. 2017;20:874–90.
Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–4.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no financial conflicts of interest.
Ethical statement
No procedures are described that need ethical approval in this manuscript.
Additional information
A correction to this article is available online at https://doi.org/10.1007/s13770-017-0111-9.
Rights and permissions
About this article
Cite this article
Patterson, B., Tanaka, Y. & Park, IH. New Advances in Human X Chromosome Status from a Developmental and Stem Cell Biology. Tissue Eng Regen Med 14, 643–652 (2017). https://doi.org/10.1007/s13770-017-0096-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13770-017-0096-4