Abstract
Purpose of Review
Expanding therapeutic targets from proteins to RNAs opens up new possibilities for neurodegenerative disorders therapeutics development. Recently, a disease-modifying antisense oligonucleotide (ASO) agent was approved for spinal muscular atrophy, suggesting ASOs will fulfill their early promise and become a significant new therapeutic category for neurodegenerative disorders.
Recent Findings
ASOs are in human subjects testing for Huntington disease, monogenic forms of amyotrophic lateral sclerosis, Alzheimer disease, myotonic dystrophy, Leber congenital amaurosis, Usher syndrome, and retinitis pigmentosum, with many more in preclinical development. Current ASO strategies encompass RNA processing modulation, and RNA target breakdown. Broad ASO mechanism categories are protein restoring versus protein lowering. Individual ASO mechanisms of action range from mutation-specific to impacting many proteins.
Summary
Current ASOs show great promise in neurodegenerative disorders. Specific ASO designs and mechanisms may be more tenable in this disease area. Preclinical development is already leveraging early knowledge from these initial clinical trials to develop novel ASO cocktails, new ASO chemical modifications, and new ASO RNA and protein targets.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Crooke ST, Lemonidis KM, Neilson L, Griffey R, Lesnik EA, Monia BP. Kinetic characteristics of Escherichia coli RNase H1: cleavage of various antisense oligonucleotide-RNA duplexes. Biochem J. 1995;312(Pt 2):599–608. https://doi.org/10.1042/bj3120599.
Gao WY, Han FS, Storm C, Egan W, Cheng YC. Phosphorothioate oligonucleotides are inhibitors of human DNA polymerases and RNase H: implications for antisense technology. Mol Pharmacol. 1992;41(2):223–9.
• Bennett CF. Therapeutic antisense oligonucleotides are coming of age. Annu Rev Med. 2019;70:307–21. https://doi.org/10.1146/annurev-med-041217-010829Overview of ASO therapeutic strategies and pharmacology focused on clinically available agents.
Scoles DR, Pulst SM. Oligonucleotide therapeutics in neurodegenerative diseases. RNA Biol. 2018;15(6):707–14. https://doi.org/10.1080/15476286.2018.1454812.
• Egli M, Manoharan M. Re-engineering RNA molecules into therapeutic agents. Acc Chem Res. 2019;52(4):1036–47. https://doi.org/10.1021/acs.accounts.8b00650Detailed look at chemical modifications and their impact on nucleic acids as therapeutics.
Crooke ST, Wang S, Vickers TA, Shen W, Liang XH. Cellular uptake and trafficking of antisense oligonucleotides. Nat Biotechnol. 2017;35(3):230–7. https://doi.org/10.1038/nbt.3779.
Geary RS, Norris D, Yu R, Bennett CF. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev. 2015;87:46–51. https://doi.org/10.1016/j.addr.2015.01.008.
Rigo F, Chun SJ, Norris DA, Hung G, Lee S, Matson J, et al. Pharmacology of a central nervous system delivered 2'-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J Pharmacol Exp Ther. 2014;350(1):46–55. https://doi.org/10.1124/jpet.113.212407.
•• Bennett CF, Baker BF, Pham N, Swayze E, Geary RS. Pharmacology of Antisense Drugs. Annu Rev Pharmacol Toxicol. 2017;57:81–105. https://doi.org/10.1146/annurev-pharmtox-010716-104846Detailed review of ASO chemical modifications, pharmacokinetics, toxicology.
Monia BP, Lesnik EA, Gonzalez C, Lima WF, McGee D, Guinosso CJ, et al. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J Biol Chem. 1993;268(19):14514–22.
Sharp PA. The centrality of RNA. Cell. 2009;136(4):577–80. https://doi.org/10.1016/j.cell.2009.02.007.
Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res. 2015;25(10):1456–65. https://doi.org/10.1101/gr.191122.115.
Bartel DP. Metazoan MicroRNAs. Cell. 2018;173(1):20–51. https://doi.org/10.1016/j.cell.2018.03.006.
Woo CJ, Maier VK, Davey R, Brennan J, Li G, Brothers J 2nd, et al. Gene activation of SMN by selective disruption of lncRNA-mediated recruitment of PRC2 for the treatment of spinal muscular atrophy. Proc Natl Acad Sci U S A. 2017;114(8):E1509–E18. https://doi.org/10.1073/pnas.1616521114.
Wu J, Sun X. Complement system and age-related macular degeneration: drugs and challenges. Drug Des Devel Ther. 2019;13:2413–25. https://doi.org/10.2147/DDDT.S206355.
Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–65. https://doi.org/10.1016/0092-8674(95)90460-3.
Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, et al. Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study. Lancet. 2016;388(10063):3017–26. https://doi.org/10.1016/S0140-6736(16)31408-8.
• Vazquez-Dominguez I, Garanto A, Collin RWJ. Molecular therapies for inherited retinal diseases-current standing, opportunities and challenges. Genes (Basel). 2019;10(9). https://doi.org/10.3390/genes10090654Overview of a key growing area in molecular therapeutics for neurodegenerative disorders.
Rowe-Rendleman CL, Durazo SA, Kompella UB, Rittenhouse KD, Di Polo A, Weiner AL, et al. Drug and gene delivery to the back of the eye: from bench to bedside. Invest Ophthalmol Vis Sci. 2014;55(4):2714–30. https://doi.org/10.1167/iovs.13-13707.
Vitravene Study G. Safety of intravitreous fomivirsen for treatment of cytomegalovirus retinitis in patients with AIDS. Am J Ophthalmol. 2002;133(4):484–98. https://doi.org/10.1016/s0002-9394(02)01332-6.
Vitravene Study G. A randomized controlled clinical trial of intravitreous fomivirsen for treatment of newly diagnosed peripheral cytomegalovirus retinitis in patients with AIDS. Am J Ophthalmol. 2002;133(4):467–74. https://doi.org/10.1016/s0002-9394(02)01327-2.
Khan M, Fadaie Z, Cornelis SS, Cremers FPM, Roosing S. Identification and analysis of genes associated with inherited retinal diseases. Methods Mol Biol. 1834;2019:3–27. https://doi.org/10.1007/978-1-4939-8669-9_1.
Hammond SM, Wood MJ. Genetic therapies for RNA mis-splicing diseases. Trends Genet. 2011;27(5):196–205. https://doi.org/10.1016/j.tig.2011.02.004.
Bacchi N, Casarosa S, Denti MA. Splicing-correcting therapeutic approaches for retinal dystrophies: where endogenous gene regulation and specificity matter. Invest Ophthalmol Vis Sci. 2014;55(5):3285–94. https://doi.org/10.1167/iovs.14-14544.
den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KE, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006;79(3):556–61. https://doi.org/10.1086/507318.
Dulla K, Aguila M, Lane A, Jovanovic K, Parfitt DA, Schulkens I, et al. Splice-modulating oligonucleotide QR-110 restores CEP290 mRNA and function in human c.2991+1655A>G LCA10 models. Mol Ther Nucleic Acids. 2018;12:730–40. https://doi.org/10.1016/j.omtn.2018.07.010.
•• Cideciyan AV, Jacobson SG, Drack AV, Ho AC, Charng J, Garafalo AV, et al. Effect of an intravitreal antisense oligonucleotide on vision in Leber congenital amaurosis due to a photoreceptor cilium defect. Nat Med. 2019;25(2):225–8. https://doi.org/10.1038/s41591-018-0295-0Although this is a small open label study, it is one of very few peer-reviewed publications of clinical trial data in this field.
Mathur P, Yang J. Usher syndrome: hearing loss, retinal degeneration and associated abnormalities. Biochim Biophys Acta. 2015;1852(3):406–20. https://doi.org/10.1016/j.bbadis.2014.11.020.
• Hastings ML, Jones TA. Antisense Oligonucleotides for the Treatment of Inner Ear Dysfunction. Neurotherapeutics. 2019;16(2):348–59. https://doi.org/10.1007/s13311-019-00729-0Overview of pros and cons of ASO therapeutics in a distinct area of neurological disease.
Yan D, Liu XZ. Genetics and pathological mechanisms of usher syndrome. J Hum Genet. 2010;55(6):327–35. https://doi.org/10.1038/jhg.2010.29.
Yan D, Ouyang X, Patterson DM, Du LL, Jacobson SG, Liu XZ. Mutation analysis in the long isoform of USH2A in American patients with usher syndrome type II. J Hum Genet. 2009;54(12):732–8. https://doi.org/10.1038/jhg.2009.107.
•• Li D, Mastaglia FL, Fletcher S, Wilton SD. Precision medicine through antisense oligonucleotide-mediated exon skipping. Trends Pharmacol Sci. 2018;39(11):982–94. https://doi.org/10.1016/j.tips.2018.09.001.detailsWork on a specific ASO strategy relevant to neurodegenerative and other neurological disorders.
Al-Chalabi A, Durr A, Wood NW, Parkinson MH, Camuzat A, Hulot JS, et al. Genetic variants of the alpha-synuclein gene SNCA are associated with multiple system atrophy. PLoS One. 2009;4(9):e7114. https://doi.org/10.1371/journal.pone.0007114.
Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62.
• Ly CV, Miller TM. Emerging antisense oligonucleotide and viral therapies for amyotrophic lateral sclerosis. Curr Opin Neurol. 2018;31(5):648–54. https://doi.org/10.1097/WCO.0000000000000594Covers clinical and pre-clinical work relevant to ALS and other neurodegenerative disorders.
Marangi G, Traynor BJ. Genetic causes of amyotrophic lateral sclerosis: new genetic analysis methodologies entailing new opportunities and challenges. Brain Res. 1607;2015:75–93. https://doi.org/10.1016/j.brainres.2014.10.009.
Miller TM, Pestronk A, David W, Rothstein J, Simpson E, Appel SH, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol. 2013;12(5):435–42. https://doi.org/10.1016/S1474-4422(13)70061-9.
McCampbell A, Cole T, Wegener AJ, Tomassy GS, Setnicka A, Farley BJ, et al. Antisense oligonucleotides extend survival and reverse decrement in muscle response in ALS models. J Clin Invest. 2018;128(8):3558–67. https://doi.org/10.1172/JCI99081.
Miller TM, Cudkowicz ME, Shaw PJ, Graham D, Fradette S, Houshyar H, et al. Safety, PK, PD, and exploratory efficacy in a single and multiple-dose study of a SOD1 antisense oligonucleotide (BIIB067) administered to participants with ALS. Neurology. 2019;92(15 Supplement):Emerging Science Abstracts. https://doi.org/10.1212/WNL.0000000000007887.
Spillantini MG, Goedert M. Tau pathology and neurodegeneration. Lancet Neurol. 2013;12(6):609–22. https://doi.org/10.1016/S1474-4422(13)70090-5.
Zhang CC, Zhu JX, Wan Y, Tan L, Wang HF, Yu JT, et al. Meta-analysis of the association between variants in MAPT and neurodegenerative diseases. Oncotarget. 2017;8(27):44994–5007. https://doi.org/10.18632/oncotarget.16690.
Schoch KM, DeVos SL, Miller RL, Chun SJ, Norrbom M, Wozniak DF, et al. Increased 4R-tau induces pathological changes in a human-tau mouse model. Neuron. 2016;90(5):941–7. https://doi.org/10.1016/j.neuron.2016.04.042.
DeVos SL, Miller RL, Schoch KM, Holmes BB, Kebodeaux CS, Wegener AJ, et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci Transl Med. 2017;9(374). https://doi.org/10.1126/scitranslmed.aag0481.
Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82(4):239–59. https://doi.org/10.1007/BF00308809.
Braak H, Thal DR, Ghebremedhin E, Del Tredici K. Stages of the pathologic process in Alzheimer disease: age categories from 1 to 100 years. J Neuropathol Exp Neurol. 2011;70(11):960–9. https://doi.org/10.1097/NEN.0b013e318232a379.
•• Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, Wild EJ, Saft C, Barker RA, et al. Targeting Huntingtin expression in patients with Huntington’s disease. N Engl J Med. 2019;380(24):2307–16. https://doi.org/10.1056/NEJMoa1900907Peer reviewed report of initial clinical trial data for an ASO in a CNS-based neurodegenerative disease.
Bates GP, Dorsey R, Gusella JF, Hayden MR, Kay C, Leavitt BR, et al. Huntington disease. Nat Rev Dis Primers. 2015;1:15005. https://doi.org/10.1038/nrdp.2015.5.
Saudou F, Humbert S. The biology of Huntingtin. Neuron. 2016;89(5):910–26. https://doi.org/10.1016/j.neuron.2016.02.003.
• Wiatr K, Szlachcic WJ, Trzeciak M, Figlerowicz M, Figiel M. Huntington Disease as a neurodevelopmental disorder and early Signs of the Disease in Stem Cells. Mol Neurobiol. 2018;55(4):3351–71. https://doi.org/10.1007/s12035-017-0477-7Important concepts to consider in non-allele specific protein lowering therapeutic strategies, even for apparently straightforward autosomal dominant mutations.
Lee JM, Ramos EM, Lee JH, Gillis T, Mysore JS, Hayden MR, et al. CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology. 2012;78(10):690–5. https://doi.org/10.1212/WNL.0b013e318249f683.
Gusella JF, MacDonald ME, Lee JM. Genetic modifiers of Huntington’s disease. Mov Disord. 2014;29(11):1359–65. https://doi.org/10.1002/mds.26001.
Squitieri F, Gellera C, Cannella M, Mariotti C, Cislaghi G, Rubinsztein DC, et al. Homozygosity for CAG mutation in Huntington disease is associated with a more severe clinical course. Brain. 2003;126(Pt 4):946–55. https://doi.org/10.1093/brain/awg077.
Ganesh A, Galetta S. Editors’ note: clinical manifestations of homozygote allele carriers in Huntington disease. Neurology. 2020;94(16):722. https://doi.org/10.1212/WNL.0000000000009305.
Cubo E, Martinez-Horta SI, Santalo FS, Descalls AM, Calvo S, Gil-Polo C, et al. Clinical manifestations of homozygote allele carriers in Huntington disease. Neurology. 2019;92(18):e2101–e8. https://doi.org/10.1212/WNL.0000000000007147.
Squitieri F, Andrew SE, Goldberg YP, Kremer B, Spence N, Zeisler J, et al. DNA haplotype analysis of Huntington disease reveals clues to the origins and mechanisms of CAG expansion and reasons for geographic variations of prevalence. Hum Mol Genet. 1994;3(12):2103–14.
Warby SC, Visscher H, Collins JA, Doty CN, Carter C, Butland SL, et al. HTT haplotypes contribute to differences in Huntington disease prevalence between Europe and East Asia. Eur J Hum Genet. 2011;19(5):561–6. https://doi.org/10.1038/ejhg.2010.229.
Kay C, Hayden MR, Leavitt BR. Epidemiology of Huntington disease. Handb Clin Neurol. 2017;144:31–46. https://doi.org/10.1016/B978-0-12-801893-4.00003-1.
Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, DiFiglia M, et al. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr Biol. 2009;19(9):774–8. https://doi.org/10.1016/j.cub.2009.03.030.
Kay C, Collins JA, Skotte NH, Southwell AL, Warby SC, Caron NS, et al. Huntingtin haplotypes provide prioritized target panels for allele-specific silencing in Huntington disease patients of European ancestry. Mol Ther. 2015;23(11):1759–71. https://doi.org/10.1038/mt.2015.128.
Castilhos RM, Augustin MC, Santos JA, Perandones C, Saraiva-Pereira ML, Jardim LB, et al. Genetic aspects of Huntington’s disease in Latin America. A systematic review. Clin Genet. 2016;89(3):295–303. https://doi.org/10.1111/cge.12641.
Baine FK, Kay C, Ketelaar ME, Collins JA, Semaka A, Doty CN, et al. Huntington disease in the South African population occurs on diverse and ethnically distinct genetic haplotypes. Eur J Hum Genet. 2013;21(10):1120–7. https://doi.org/10.1038/ejhg.2013.2.
•• Kay C, Collins JA, Caron NS, Agostinho LA, Findlay-Black H, Casal L, et al. A comprehensive haplotype-targeting strategy for Aallele-specific HTT suppression in Huntington disease. Am J Hum Genet. 2019;105(6):1112–25. https://doi.org/10.1016/j.ajhg.2019.10.011. Key considerations in any haplotype-dependent ASO therapeutic strategy.
Pulkes T, Papsing C, Wattanapokayakit S, Mahasirimongkol S. CAG-expansion haplotype analysis in a population with a low prevalence of Huntington’s disease. J Clin Neurol. 2014;10(1):32–6. https://doi.org/10.3988/jcn.2014.10.1.32.
Wild EJ, Tabrizi SJ. Therapies targeting DNA and RNA in Huntington’s disease. Lancet Neurol. 2017;16(10):837–47. https://doi.org/10.1016/S1474-4422(17)30280-6.
Iwamoto N, Butler DCD, Svrzikapa N, Mohapatra S, Zlatev I, Sah DWY, et al. Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides. Nat Biotechnol. 2017;35(9):845–51. https://doi.org/10.1038/nbt.3948.
Dryja TP, McGee TL, Reichel E, Hahn LB, Cowley GS, Yandell DW, et al. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature. 1990;343(6256):364–6. https://doi.org/10.1038/343364a0.
Sullivan LS, Bowne SJ, Birch DG, Hughbanks-Wheaton D, Heckenlively JR, Lewis RA, et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest Ophthalmol Vis Sci. 2006;47(7):3052–64. https://doi.org/10.1167/iovs.05-1443.
Talib M, Boon CJF. Retinal dystrophies and the road to treatment: clinical requirements and considerations. Asia Pac J Ophthalmol (Phila). 2020;9(3):159–79. https://doi.org/10.1097/APO.0000000000000290.
Murray SF, Jazayeri A, Matthes MT, Yasumura D, Yang H, Peralta R, et al. Allele-specific inhibition of rhodopsin with an antisense oligonucleotide slows photoreceptor cell degeneration. Invest Ophthalmol Vis Sci. 2015;56(11):6362–75. https://doi.org/10.1167/iovs.15-16400.
•• Chen KW, Chen JA. Functional roles of long non-coding RNAs in motor neuron development and disease. J Biomed Sci. 2020;27(1):38. https://doi.org/10.1186/s12929-020-00628-zKey concepts relevant to ASO therapeutics development across lncRNA-based neurological disorders.
Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann Neurol. 2010;67(3):291–300. https://doi.org/10.1002/ana.21948.
Thornton CA. Myotonic dystrophy. Neurol Clin. 2014;32(3):705–19, viii. https://doi.org/10.1016/j.ncl.2014.04.011.
Meola G, Cardani R. Myotonic dystrophies: an update on clinical aspects, genetic, pathology, and molecular pathomechanisms. Biochim Biophys Acta. 2015;1852(4):594–606. https://doi.org/10.1016/j.bbadis.2014.05.019.
•• Thornton CA, Wang E, Carrell EM. Myotonic dystrophy: approach to therapy. Curr Opin Genet Dev. 2017;44:135–40. https://doi.org/10.1016/j.gde.2017.03.007Key concepts relevant to ASO therapeutics design across lncRNA-based neurological disorders, particularly multisystem disorders and disorders where lncRNA pathology has wide-ranging primary and secondary impacts.
Musova Z, Mazanec R, Krepelova A, Ehler E, Vales J, Jaklova R, et al. Highly unstable sequence interruptions of the CTG repeat in the myotonic dystrophy gene. Am J Med Genet A. 2009;149A(7):1365–74. https://doi.org/10.1002/ajmg.a.32987.
Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;69(2):385. https://doi.org/10.1016/0092-8674(92)90418-c.
Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488(7409):111–5. https://doi.org/10.1038/nature11362.
Pandey SK, Wheeler TM, Justice SL, Kim A, Younis HS, Gattis D, et al. Identification and characterization of modified antisense oligonucleotides targeting DMPK in mice and nonhuman primates for the treatment of myotonic dystrophy type 1. J Pharmacol Exp Ther. 2015;355(2):329–40. https://doi.org/10.1124/jpet.115.226969.
Mignon L. https://www.myotonic.org/sites/default/files/pages/files/Laurence-Mignon_IONIS-Update_2018-Conference.pdf. Myotonic Dystrophy Foundation meeting 2018.
Yadava RS, Yu Q, Mandal M, Rigo F, Bennett CF, Mahadevan MS. Systemic therapy in a RNA toxicity mouse model with an antisense oligonucleotide therapy targeting a non-CUG sequence within the DMPK 3'UTR RNA. Hum Mol Genet. 2020;29:1440–53. https://doi.org/10.1093/hmg/ddaa060.
Klein AF, Varela MA, Arandel L, Holland A, Naouar N, Arzumanov A, et al. Peptide-conjugated oligonucleotides evoke long-lasting myotonic dystrophy correction in patient-derived cells and mice. J Clin Invest. 2019;129(11):4739–44. https://doi.org/10.1172/JCI128205.
Christou M, Wengel J, Sokratous K, Kyriacou K, Nikolaou G, Phylactou LA, et al. Systemic evaluation of chimeric LNA/2'-O-methyl steric blockers for myotonic dystrophy type 1 therapy. Nucleic Acid Ther. 2020;30(2):80–93. https://doi.org/10.1089/nat.2019.0811.
Stepniak-Konieczna E, Konieczny P, Cywoniuk P, Dluzewska J, Sobczak K. AON-induced splice-switching and DMPK pre-mRNA degradation as potential therapeutic approaches for myotonic dystrophy type 1. Nucleic Acids Res. 2020;48(5):2531–43. https://doi.org/10.1093/nar/gkaa007.
Jamal GA, Weir AI, Hansen S, Ballantyne JP. Myotonic dystrophy. A reassessment by conventional and more recently introduced neurophysiological techniques. Brain. 1986;109(Pt 6):1279–96. https://doi.org/10.1093/brain/109.6.1279.
Labayru G, Diez I, Sepulcre J, Fernandez E, Zulaica M, Cortes JM, et al. Regional brain atrophy in gray and white matter is associated with cognitive impairment in Myotonic dystrophy type 1. Neuroimage Clin. 2019;24:102078. https://doi.org/10.1016/j.nicl.2019.102078.
Abramzon YA, Fratta P, Traynor BJ, Chia R. The overlapping genetics of amyotrophic lateral sclerosis and frontotemporal dementia. Front Neurosci. 2020;14:42. https://doi.org/10.3389/fnins.2020.00042.
Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79(3):416–38. https://doi.org/10.1016/j.neuron.2013.07.033.
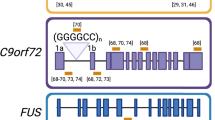
DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–56. https://doi.org/10.1016/j.neuron.2011.09.011.
Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–68. https://doi.org/10.1016/j.neuron.2011.09.010.
Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11(4):323–30. https://doi.org/10.1016/S1474-4422(12)70043-1.
Rademakers R. C9orf72 repeat expansions in patients with ALS and FTD. Lancet Neurol. 2012;11(4):297–8. https://doi.org/10.1016/S1474-4422(12)70046-7.
Haeusler AR, Donnelly CJ, Rothstein JD. The expanding biology of the C9orf72 nucleotide repeat expansion in neurodegenerative disease. Nat Rev Neurosci. 2016;17(6):383–95. https://doi.org/10.1038/nrn.2016.38.
Cappella M, Ciotti C, Cohen-Tannoudji M, Biferi MG. Gene therapy for ALS-a perspective. Int J Mol Sci. 2019;20(18). https://doi.org/10.3390/ijms20184388.
Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A. 2013;110(47):E4530–9. https://doi.org/10.1073/pnas.1318835110.
Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, Seelman A, et al. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron. 2016;90(3):535–50. https://doi.org/10.1016/j.neuron.2016.04.006.
Frazier KS. Antisense oligonucleotide therapies: the promise and the challenges from a toxicologic pathologist’s perspective. Toxicol Pathol. 2015;43(1):78–89. https://doi.org/10.1177/0192623314551840.
Aupy P, Echevarria L, Relizani K, Zarrouki F, Haeberli A, Komisarski M, et al. Identifying and avoiding tcDNA-ASO sequence-specific toxicity for the development of DMD exon 51 skipping therapy. Mol Ther Nucleic Acids. 2020;19:371–83. https://doi.org/10.1016/j.omtn.2019.11.020.
Crooke ST, Baker BF, Witztum JL, Kwoh TJ, Pham NC, Salgado N, et al. The effects of 2'-O-Methoxyethyl containing antisense oligonucleotides on platelets in human clinical trials. Nucleic Acid Ther. 2017;27(3):121–9. https://doi.org/10.1089/nat.2016.0650.
Schoch KM, Miller TM. Antisense oligonucleotides: translation from mouse models to human neurodegenerative diseases. Neuron. 2017;94(6):1056–70. https://doi.org/10.1016/j.neuron.2017.04.010.
Wild EJ, Tabrizi SJ. Targets for future clinical trials in Huntington’s disease: what’s in the pipeline? Mov Disord. 2014;29(11):1434–45. https://doi.org/10.1002/mds.26007.
Ammala C, Drury WJ 3rd, Knerr L, Ahlstedt I, Stillemark-Billton P, Wennberg-Huldt C, et al. Targeted delivery of antisense oligonucleotides to pancreatic beta-cells. Sci Adv. 2018;4(10):eaat3386. https://doi.org/10.1126/sciadv.aat3386.
Huang Y. Preclinical and clinical advances of GalNAc-decorated nucleic acid therapeutics. Mol Ther Nucleic Acids. 2017;6:116–32. https://doi.org/10.1016/j.omtn.2016.12.003.
Tsimikas S, Viney NJ, Hughes SG, Singleton W, Graham MJ, Baker BF, et al. Antisense therapy targeting apolipoprotein(a): a randomised, double-blind, placebo-controlled phase 1 study. Lancet. 2015;386(10002):1472–83. https://doi.org/10.1016/S0140-6736(15)61252-1.
Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, Bondulich MK, et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci U S A. 2013;110(6):2366–70. https://doi.org/10.1073/pnas.1221891110.
Michel L, Huguet-Lachon A, Gourdon G. Sense and antisense DMPK RNA foci accumulate in DM1 tissues during development. PLoS One. 2015;10(9):e0137620. https://doi.org/10.1371/journal.pone.0137620.
De Vivo DC, Bertini E, Swoboda KJ, Hwu WL, Crawford TO, Finkel RS, et al. Nusinersen initiated in infants during the presymptomatic stage of spinal muscular atrophy: interim efficacy and safety results from the phase 2 NURTURE study. Neuromuscul Disord. 2019;29(11):842–56. https://doi.org/10.1016/j.nmd.2019.09.007.
• Testa CM, Jankovic J. Huntington disease: a quarter century of progress since the gene discovery. J Neurol Sci. 2019;396:52–68. https://doi.org/10.1016/j.jns.2018.09.022Includes framework for understanding challenges for early therapeutic intervention concepts, particularly for neurodegenerative disorders with a wide phenotype range.
• Aartsma-Rus A, Straub V, Hemmings R, Haas M, Schlosser-Weber G, Stoyanova-Beninska V, et al. Development of exon skipping therapies for Duchenne muscular dystrophy: a critical review and a perspective on the outstanding issues. Nucleic Acid Ther. 2017;27(5):251–9. https://doi.org/10.1089/nat.2017.0682Covers key ASO therapeutics challenges around treating the full patient population in a multi-etiology disorder, such as use of ASO cocktails within one therapeutic.
Rodrigues FB, Quinn L, Wild EJ. Huntington’s disease clinical trials corner: January 2019. J Huntingtons Dis. 2019;8(1):115–25. https://doi.org/10.3233/JHD-190001.
Bali T, Self W, Liu J, Siddique T, Wang LH, Bird TD, et al. Defining SOD1 ALS natural history to guide therapeutic clinical trial design. J Neurol Neurosurg Psychiatry. 2017;88(2):99–105. https://doi.org/10.1136/jnnp-2016-313521.
Hubers A, Just W, Rosenbohm A, Muller K, Marroquin N, Goebel I, et al. De novo FUS mutations are the most frequent genetic cause in early-onset German ALS patients. Neurobiol Aging. 2015;36(11):3117 e1-e6. https://doi.org/10.1016/j.neurobiolaging.2015.08.005.
Conte A, Lattante S, Zollino M, Marangi G, Luigetti M, Del Grande A, et al. P525L FUS mutation is consistently associated with a severe form of juvenile amyotrophic lateral sclerosis. Neuromuscul Disord. 2012;22(1):73–5. https://doi.org/10.1016/j.nmd.2011.08.003.
Deusterward R. https://patientworthy.com/2019/06/10/the-fda-congress-a-young-woman-dying-of-als-her-physician-and-her-parents-are-all-struggling-over-access-to-an-untested-therapy/. 2019.
Figueiredo M. https://alsnewstoday.com/2020/03/16/jacifusen-collaboration-funds-experimental-therapy-for-patients-with-rare-fus-als/. ALS News Today. 2020.
Thakur N. https://www.als.org/stories-news/deeper-look-als-association-efforts-speed-approval-gene-therapies. 2020.
• Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–22. https://doi.org/10.1038/nrd.2016.246A look at a potential new ASO therapeutic class.
d'Ydewalle C, Ramos DM, Pyles NJ, Ng SY, Gorz M, Pilato CM, et al. The antisense transcript SMN-AS1 regulates SMN expression and is a novel therapeutic target for spinal muscular atrophy. Neuron. 2017;93(1):66–79. https://doi.org/10.1016/j.neuron.2016.11.033.
Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, et al. Stress granule assembly disrupts nucleocytoplasmic transport. Cell. 2018;173(4):958–71 e17. https://doi.org/10.1016/j.cell.2018.03.025.
Becker LA, Huang B, Bieri G, Ma R, Knowles DA, Jafar-Nejad P, et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature. 2017;544(7650):367–71. https://doi.org/10.1038/nature22038.
Carroll JB, Bates GP, Steffan J, Saft C, Tabrizi SJ. Treating the whole body in Huntington’s disease. Lancet Neurol. 2015;14(11):1135–42. https://doi.org/10.1016/S1474-4422(15)00177-5.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Neurology of Aging
Rights and permissions
About this article
Cite this article
Testa, C.M. Antisense Oligonucleotide Therapeutics for Neurodegenerative Disorders. Curr Geri Rep 11, 19–32 (2022). https://doi.org/10.1007/s13670-020-00341-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13670-020-00341-7