Abstract
There is growing interest in selective breeding of the honeybee, resulting in the emergence of new breeding projects, often with an emphasis on improving resilience traits, in particular toward brood diseases. Lately, feed autonomy is also gaining importance. Here, we use data from a small breeding nucleus in France to estimate genetic parameters for common bee breeding traits and a novel trait reflecting honey reserves in the brood chamber. Open-mated queens were produced each year from inseminated dams between 2019 and 2021, and ~330 colonies were phenotyped each following year at three periods during the entire beekeeping season. Genetic parameters were estimated using ReML with an animal model. Narrow-sense heritability estimates ranged from low (around 0.15) for calmness and total capped brood surface both measured in early summer, to moderate (0.30 to 0.40) for hygienic behavior in spring, honey yield, and phoretic V. destructor load in early summer. Honey reserves in the brood chamber showed an intermediate heritability throughout the season (around 0.25). Gentleness had a null heritability. Most correlations between phenotypes adjusted for environmental fixed effects were close to zero. Among exceptions, there were honey reserves in the brood chamber in early summer with honey yield (around −0.2) and with the total capped brood surface in early summer (around −0.3). These estimates, although uncertain due to the dataset size, suggest that selection for production and resilience will be effective, even though simultaneous selection for honey yield and feed reserves might be difficult due to a possible genetic antagonism between both traits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Worldwide, there are few organized breeding programs for honeybees (Apis mellifera), with a few notable exceptions, such as in Austria and Germany (Rinderer et al. 2010; Danka et al. 2016; Brascamp et al. 2016; Hoppe et al. 2020). Nevertheless, there is growing interest in selective breeding of the species, resulting in the emergence of new breeding projects. Engaging in selective breeding can enable a group of beekeepers to maintain and develop original stock well adapted to their specific management practices. Breeding focuses usually on enhancing classical traits such as honey production, handling ease, and reducing the tendency to swarm.
In addition to these classical selection objectives, there is a strong emphasis on improving traits related to resilience of the colonies. In farm animals, resilience is defined as the capacity of animals (here colonies) to remain productive when exposed to environmental or infectious challenges (Colditz and Hine 2016). Part of the effort to improve resilience is linked to disease resistance, including via the control of the parasitic V. destructor mite (Rinderer et al. 2010; Danka et al. 2016; Büchler et al. 2020; Guichard et al. 2020a). Furthermore, there is also a growing interest in enhancing feed autonomy of colonies when facing fluctuating feed resources, as feed shortages are common in a wide range of landscapes (e.g., Czekońska et al. 2023). In such cases, feed autonomy is the capacity of a colony to survive and become productive again as soon as feed shortage is over, without needing emergency sugar feeding. Concern over feed autonomy may further be exacerbated by climate change and the reduced diversity of melliferous plants in agricultural landscapes (Decourtye et al. 2010; Goulson et al. 2015). Other reasons for considering feed autonomy as a breeding goal trait include the general intensification of beekeeping (e.g., more fecund stock, several harvests per season); the increase in sugar prices; and stricter controls on honey purity by international dealers that might detect trace amounts of emergency feeding, trying to avoid buying adulterated honey (Ždiniaková et al. 2023).
In the context of emerging breeding schemes and new traits, colony performance records and associated pedigrees are only available on relatively small populations with few generations of trait recording (e.g., Guichard et al. 2020b). Moreover, important traits are usually thought to arise genetically from effects jointly expressed by the queen and its worker group. In consequence, the colony phenotype is usually described by a mixed model containing two random genetic effects: an effect expressed by the queen and an effect of the worker group (Chevalet and Cornuet 1982; Bienefeld et al. 1989). We will refer to this model as the Colony Model (CM). Distinguishing between the genetic effects of the queen and of the workers on colony performance, however, is only possible when the two castes have distinct pedigrees and the dataset is large enough. In datasets from small populations, limited pedigree depth, or containing only records from colonies with open-mated queens that are not used to produce queens for subsequent generations, the CM cannot be fitted. To still obtain information on the heritability of traits, reduced models can be used that include only the genetic effect of either the queen or the worker group of the colony (Guichard et al. 2020b; Du et al. 2022; Basso et al. 2024).
Using a reduced model with only a queen effect (Queen Model: QM), we will present genetic parameter estimates on a small population from an emerging breeding scheme managed by the Association for the Development of Beekeeping in the Provence region (ADAPI) in southern France. Their breeding objective focuses on improving honey yield and resilience to diseases and to fluctuating feed resources, for which a new selection criterion was developed.
2 Material and methods
2.1 Breeding population
During the first 3 years of initialization of the breeding program (2019–2021), the population under study was of Buckfast origin (a synthetic breed), partially hybridized with Caucasian and dark honeybees (for a description of honeybee subspecies or breeds, see Adam (1987) and Ruttner (2013)). Founders were chosen primarily for good honey production, feed autonomy, or V. destructor resilience.
From 2019 to 2021, 14 to 16 breeding queens (BQs) (Table I) were used each year to produce offspring queen families. In total, there were 42 BQs, from which 26 were used to produce offspring queens tested on several apiaries, while the others were private BQs used to produce an offspring sister-group tested in a single apiary.
Genetic links from one generation to the other existed through the dam and sire path and are detailed in Supplementary Figure S1.
Two types of daughters were produced by BQs. The first one was composed of potential new BQs that were inseminated. To allow for a short generation interval, BQs were not phenotyped or selected. The second one was composed of open-mated daughters. Shared open-mated daughters were produced by five (in 2019) or four (in 2020 and 2021) queen producers and mated freely with drones from the area of each queen producer. They were then distributed to the performance-testing beekeepers and will be referred to as shared Testing Queens (TQs). In addition to these shared TQ sister-groups, private TQ sister-groups (i.e., not shared between beekeepers) were open-mated and tested by their owners. TQs from a dam used in 2020 were used as drone-producing queens for the insemination of BQs used in 2021 (see Supplementary Figure S1 for details). All other TQs were only phenotyped and not used for mating.
2.2 Testing protocol
Each year from 2020 to 2022, 13 to 15 beekeepers managed their own testing apiary using the same testing protocol and otherwise maintained their normal production management. Colonies were migratory, except those managed by one of the beekeepers in 2021. Each apiary generally tested three groups of shared TQ sister-groups or at least two. Each shared TQ sister-group was tested on three to five apiaries to ensure a sufficient genetic connection between apiaries. In addition to the shared TQ sister-groups, around half of the beekeepers tested a private TQ sister-group, present only in their own apiary. TQs were introduced in homogenized queen-less colonies to be performance-tested in the following season (Büchler et al. 2013). After winter mortality, requeening, swarming, or colony collapse due to diseases, a total of 1022 colonies remained with phenotypes over the whole beekeeping season, with a minimum of 330 per year (Table I).
Phenotyping during the production season started at the end of winter, in March–April, and ended in mid-summer (mid-August) after the lavender bloom, the main targeted honey flow. Honey yield (HONEY) was measured as the sum of all honey harvested from supers during the whole season. Colonies that produced zero kg of honey but otherwise showed no obvious disease symptoms, requeening, swarming, or other accidental events were considered genetically informative and kept in the analysis.
To record traits and collect samples, trained operators visited all colonies during three periods per year: end of winter (March to April), spring (late April to May), and early summer (June to mid-July). Six traits were recorded at least two times in a year. We used the phenotyping protocols described in Büchler et al. (2013) for the common traits gentleness (GENT), calmness during inspection (CALM), hygienic behavior (HYG), and phoretic V. destructor load (VARROA), with some modifications of the protocols for HYG and VARROA explained in the following paragraph. In addition, we visually assessed the total capped worker brood surface (BROOD) using the protocol defined by Hernandez et al. (2020). The protocol for the newly introduced trait representing honey reserves in the brood chamber (RESERVES) is detailed in Appendix A and aims at evaluating the quantity of honey stored in the brood chamber on a one-to-four scale. As these feed reserves are not harvested, unlike those stored in the supers, colonies could rely on them during honey flow shortages. This new trait was used to assess the potential feed autonomy of the colonies.
For HYG, 50 or 100 capped brood cells in a patch with white to purple-eyed nymphs were pierced with an entomological needle (0.45 mm in diameter) before replacing it in the colony. After a waiting time, the pinned brood cells with partially removed brood were summed together with those totally cleaned to calculate the clearance rate numerator, as, e.g., in Eynard et al. (2020). To enable a complete phenotyping of an apiary in a single visit, waiting time between pin-killing and brood removal measurement was shortened from 12 or 24 to 5 h at the end of winter and to 3.5 h in spring compared to usual recommendations (Büchler et al. 2013).
For VARROA, around 30 g samples of adult worker bees per colony and per visit were collected on open brood combs and then frozen, weighted, and washed with soapy water to detach mites from the workers (Dietemann et al. 2013). The phoretic V. destructor load was expressed per 100 bees by considering the average weight of a worker to be 0.14 g. A first infestation rate was estimated at the end of winter on all colonies to verify the initial absence of detectable mites.
In addition to these phenotypes, the presence of the desired queens (marked) was checked at each visit to ensure no requeening event had occurred. A last thorough checking was done in mid-August to verify the presence of all queens.
All traits were measured at the end of winter (except for BROOD and VARROA), spring, and early summer (except for HYG). All traits were measured each year except for HYG and RESERVES, which were not measured in 2020. Furthermore, no phenotypes were collected at the end of winter in 2020. Lastly, VARROA was not measured in spring in 2020.
2.3 Pedigree information
The pedigree included the known ancestors of BQs, the BQs themselves, and their offspring TQs. All BQs were inseminated. Each entry of the pedigree file corresponded to a queen. Each line contained the identification of the queen, the queen’s dam, and the queen’s mate (pseudo sire). Further columns characterized the mate of the queen (a pseudo sire) by the identification of the dam of the drone-producing queens (DPQs), the group of DPQs with their expected group size, and lastly, the expected number of drones (ND) it contributed to the mating of the queen.
For BQs, however, only the dam of DPQs used during instrumental insemination was recorded, but not which specific DPQs. To complete the pedigree and in accordance with the beekeepers’ insemination practice, we assumed that one group of three (NS = 3) sister-DPQs was used per insemination and that, for a particular dam of DPQs, this sister group was the same for inseminations performed in a year. According to Kistler et al. (2024), this assumption minimizes the probability of strongly deviating estimates of genetic parameters when the number of DPQ(s) used is uncertain and lies between one and three DPQs. The number of drones used for each artificial insemination was set to ND = 8.
For the open-mated TQs, we hypothesized that a dummy pseudo sire composed of a large (NS = 100) number of unrelated DPQs of unknown origin contributed ND = 12 drones for each open mating to model natural mating conditions (Baudry et al. 1998; Tarpy and Nielsen 2002; Schlüns et al. 2005; Tarpy et al. 2013). Setting the number of DPQs making up this dummy open mating pseudo sire to 100 meant that two TQs’ worker groups were considered paternally almost unrelated.
The pedigree included a total of 1087 colonies, including 1022 TQs, 42 dams (of which 20 are founders) used to produce TQ sister-groups (shared and private), and 23 maternal or paternal ancestors.
2.4 Statistical analysis
The pedigree was used to compute the inversed relationship matrix \(\mathbf {{A}^{-1}}\) following Brascamp and Bijma (2014, 2019).
The performance file contained one entry for each of the 1022 TQ colonies, with identification of the queen, the worker group, the traits’ records, the contemporary group, and the operator measuring each trait at each period. The contemporary group was defined as the interaction between the testing year, apiary, and the queen producer who bred the TQs. Pairwise Pearson’s correlation coefficients were calculated between all phenotypic values, considering all pair-wise complete observations.
To estimate genetic parameters, the vector of phenotypes y was described using a linear mixed model with only queen effects as genetic effects (QM) (Guichard et al. 2020b; Du et al. 2022):
where b is the vector of fixed effects (general mean, CG, and operator effect when present) with a corresponding incidence matrix \(\mathbf{X\,{a}_{q}}\), is the vector of genetic effects of the queens with incidence matrix \(\mathbf{{Z}_{{q}}}\), and e is the vector of residuals. A fixed operator effect was included for HYG, CALM, GENT, BROOD, and RESERVES, as preliminary analyses revealed it had a significant effect. It was not included for HONEY nor VARROA, as the same operator for each trait always measured a complete apiary at the same period. The performance file and the inversed relationship matrix were used to estimate genetic parameters and solve the BLUP equations (Henderson 1973) using AIREMLF90 (ver. 1.149) of the BLUPF90 package (Misztal et al. 2002).
All estimations were first run in single trait analyses. The heritability \(\mathbf{{h}_{{QM}}^{2}}\) was defined as the fraction of the estimated genetic variance in the QM over the phenotypic variance (sum of the genetic and residual variances of the QM). In addition to the single trait analyses, for all traits measured in early summer with a heritability estimate greater than its standard error, as well as HONEY (total seasonal yield) and HYG in spring, two-trait analyses were also run to obtain genetic correlation estimates. These correlations were compared with Pearson correlation coefficients between raw phenotypes or those adjusted for fixed effects as estimated in the BLUP model. The latter might be more informative when genetic correlation estimates are uncertain (Cheverud 1988; Roff 1995).
We performed separate single trait analyses for the same trait measured at multiple periods within the season. We did not average these measurements per colony, because genetically it may concern different traits. In particular, behavioral traits might be influenced by the sequence of measurements, with colonies possibly getting used to being visited. In addition, correction for operator effect becomes complex since operators differed for repeated records. Our data size did not allow for more sophisticated multivariate analyses.
We used R (v4.1.2; R Core Team 2021) with several packages for data formatting (tidyverse: Wickham et al. 2019; data.table: Barrett et al. 2023), computation of summary statistics (pastecs: Grosjean et al. 2018), and production of figures (corrplot: Wei and Simko 2021; rvg: Gohel 2024; officer: Gohel and Moog 2024).
3 Results
3.1 Raw phenotypes
Distributions of all traits in all periods are shown in Supplementary Figures S2 to S8.
Table II shows the descriptive statistics for all traits in all periods.
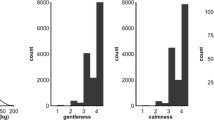
Raw coefficients of variation (CV) were highest (≫ 1.00) for both VARROA measures, followed by HYG in spring (0.95) and HONEY (0.81) measures. RESERVES and BROOD had intermediate coefficients of variation (~0.31), while GENT and CALM had the lowest ones (~0.17). Adjusting phenotypes for fixed effects reduced the CVs by, depending on the trait, 14% (for HYG at the end of winter) to 37% (for HONEY).
3.2 Genetic parameter estimates in single trait analyses
Estimated genetic parameters for all traits are shown in Table III. All estimates showed high standard errors due to the small size of the dataset. For GENT measured at any period, estimated h2 were zero. Estimated h2 were near zero also for BROOD in spring, and for CALM at the end of winter and in spring, and were far from statistically different from zero.
Traits showing h2 estimates exceeding their standard error were HONEY, CALM in early summer, BROOD in early summer, both VARROA measures, both HYG measures, and all three measures of RESERVES. Heritability estimates for these traits ranged from low (around 0.15) for CALM in early summer, BROOD in early summer, and VARROA in spring, to moderate values (0.30 to 0.40) for HYG in Spring, HONEY, and VARROA in early summer. The trait RESERVES showed an intermediate h2, ranging from 0.19 ± 0.16 at the end of winter to 0.25 ± 0.18 in early summer, as did HYG at the end of winter (0.25 ± 0.19).
3.3 Correlations between traits
Figure 1 shows the phenotypic correlations adjusted for fixed effects for all traits. Using the adjusted phenotypes did not affect Pearson’s correlations strongly compared to using raw phenotypes (Supplementary Table S1). Most phenotypic correlations were close to zero, except for some measures of the same trait between periods, such as VARROA in spring and early summer at 0.4. Other exceptions were positive correlations around 0.5 between GENT and CALM measured at the same period; RESERVES in early summer with HONEY (around −0.2) and with BROOD in early summer (around −0.3); and lastly, BROOD in spring and early summer and HONEY (around 0.3).
Correlations between phenotypes adjusted for fixed effects. Traits: HONEY, total annual honey yield (kg); GENT, gentleness (1–4 rating); CALM, calmness (1–4 rating); BROOD, capped brood surface (× 1000 cell count); VARROA, phoretic V. destructor load (mites/100 worker bees); HYG, hygienic behavior (% of totally and partially cleared cells); RESERVES, honey reserves (1–4 rating). Periods: EndWinter, end of winter (March–April); Spring (late April–May); EarlySummer, early summer (end of June–July).
We estimated genetic correlations only for traits where the h2-estimate exceeded its SE. Resulting estimates showed very large SE and are in Supplementary Table S2. These genetic correlation estimates had the same sign as the corresponding phenotypic correlations that exceeded 0.15 in absolute value.
4 Discussion
We derived estimates of heritability for usual beekeeping traits and traits related to resilience, including a new trait linked to feed autonomy during the beekeeping season, in a starting breeding program. The data included only colonies of open-mated queens without queen daughters. Because of limited data, we estimated heritability values with a Queen Model. We also clarify how our estimates relate to genetic parameters of a Colony Model. Results showed that there was sufficient genetic variability on most classical traits and on traits related to resilience to enable meaningful genetic progress, including on a new trait measuring feed reserves.
4.1 Reduced models in datasets containing only records from open-mated potential DPQs
We chose to use a reduced model (with only a genetic queen effect) because our records were all from colonies with open-mated queens (TQs) that did not contribute diploid offspring to the breeding population. Primarily because of technical limitations in this initialization phase of the breeding program, the beekeepers were not yet able to performance test sufficient BQs, but only open-mated descendants. These offspring were candidates for selection as DPQs. Open mating meant that the worker group from a tested colony always had an unknown sire. Two worker groups were therefore related only if their dams (TQs) were related, and the relationship between the worker groups fully depended on the relatedness between their dams. Suppose two tested colonies had queens related by a coefficient \(r\), then their worker groups were related by \(\frac{1}{4}r\). This made the separate contribution of queens and worker groups to colony phenotypes statistically indistinguishable. Given our data structure, the genetic variance estimated in the Queen Model (\({\widehat{\sigma }}_{{\text{A}}^{(\text{QM})}}^{2}\)) captures a quarter of the genetic variance for the worker effect (\({\sigma }_{{\text{A}}^{\text{W}}}^{2}\)), the full genetic variance for the queen effect (\({\sigma }_{{\text{A}}^{\text{Q}}}^{2}\)), and the covariance between both effects (\({\sigma }_{{\text{A}}^{\text{QW}}}\)) as defined in the Colony Model (CM). Appendix B shows the relevant mathematics, including a proof derived independently by Manuel Du (personal communication; also see Du et al. 2022, Eq. 4a).
Our data structure differs, for example, from Andonov et al.’s case (2019), where records were also from colonies with open-mated queens, but the queens were used as BQs. In such a situation, results obtained with a Queen Model (QM) do not predict results obtained with a Worker Model (WM), as pairs of colonies with records do not always have the same proportionality factor between their queen’s relationship and their worker groups’ relationship. In such cases, queen effects can potentially be distinguished from worker effects, thereby also potentially enabling the use of a Colony Model (CM).
In our specific case, QM and WM estimates could be predicted from one another because they captured the same covariance structures. However, there is no general relationship between estimates of the reduced models and the CM, nor between the two reduced models, because their estimates depend on the data structure.
4.2 Heritability estimates and comparison with the literature
HONEY and HYG showed heritability estimates in the range of what has been reported in genetic analyses using ReML parameter estimates (Brascamp et al. 2016, 2018; Andonov et al. 2019; Guichard et al. 2020b; Hoppe et al. 2020). A detailed comparison is not useful given our large SE.
Although GENT and CALM still showed phenotypic variability after correction for fixed effects, their heritability estimates were virtually zero (except CALM in early summer). This is similar to values reported for Swiss carnica (Guichard et al. 2020b). Both traits appeared heritable, however, in Swiss mellifera (Guichard et al. 2020b), and German (Hoppe et al. 2020) and Austrian (Brascamp et al. 2018) carnica.
The trait VARROA was derived from the mite count on a bee sample and, therefore, from a meristic trait with a small mean, especially in spring, where 63% of the colonies had zero mites. It had a low h2 in spring (0.13 ± 0.10). Later in the season, the proportion of colonies with zero mites decreased to 26%, increasing the mean of the trait. The h2 in early summer also appeared higher (0.33 ± 0.16). However, with the four-fold increase in the genetic standard deviation being accompanied by a proportional increase in the mean, the genetic CVs of the trait were about the same (around 0.7) in both periods.
VARROA is generally reported of low heritability, although most estimates rely on extremely small sample sizes (reviewed by Guichard et al. 2020a). Although the SE of our heritability estimates were large, VARROA appeared suited for selection with the largest genetic coefficient of variability, giving good prospects for evolvability (Houle 1992) of this trait.
We calculated the HYG score by including partially removed brood, as very few colonies had some completely removed cells within a few hours following brood damage. Furthermore, preliminary results (on 132 colonies) showed that, while early uncapped brood cells could be recapped later by workers, partially removed brood was practically always followed by complete removal after 24 h. By waiting only 3.5 h in spring, compared to 5 h in early winter, the average HYG score was further below the ideal 50% value compared with what we obtained at the end of winter, and a fifth of colonies had less than 5% cells with partially or totally removed brood. Even so, both h2 and the genetic CV for HYG seemed higher in spring than at the end of winter, although estimates were not statistically different.
The new trait RESERVES had estimates of heritability and evolvability that suggest a meaningful variability of this trait whenever it was measured from the end of winter to early summer. Comparison with more labor-intensive measurements, using the brood chamber’s weight or the visual assessment of the total feed reserves’ surface (Hernandez et al. 2020) showed that the resulting Spearman’s correlation coefficients were very high (unpubl. data). As the testing protocol (Appendix A) is relatively simple to implement, this trait could be widely used by beekeepers for their selection. Feed autonomy during the season results from the colony foraging more feed than it consumes. The capacity of foraging thus makes it another trait than feed autonomy during winter. Whether a genetic increase of the feed reserves in the brood chamber is desirable will depend heavily on managing practices (total size of the brood chamber, sedentary or moving apiaries…) and floral abundance and variability through time. Too many feed reserves in the brood chamber would eventually limit the space available for brood and result in a reduced foraging capacity. Beekeepers would probably aim at an optimum for the RESERVES trait, reached when the feed reserves stored in the brood chamber are sufficient to enable a good colony development and never necessitate emergency feeding.
Feed autonomy is expected to be favorably linked with survival. Survival in given environmental conditions is an ultimate resilience trait (i.e., fitness assessment) in animal breeding because it measures an individual’s resistance to multiple mortality factors in a specific environment. We did not study colony survival, however, because our limited dataset size could not allow for a meaningful genetic analysis of such a fitness trait that is well-known to be weakly heritable both in farm and wild animal populations due to unidentified environmental effects leading to high residual variation (Houle 1992). We may note that even from the large data set in the BeeBreed program (Bienefeld and Hoppe 2024), no estimates of genetic parameters for survival were published.
4.3 Correlations between traits
We found a phenotypic correlation of around 0.5 between GENT and CALM measured at the same period, similar to results of Hoppe et al. (2020) in German carnica. We also found a moderate positive phenotypic correlation between BROOD and HONEY, as reported in the literature (e.g., Kretzschmar and Maisonnasse 2022).
Furthermore, we found a moderate negative phenotypic correlation between BROOD and RESERVES, also reflected in the negative genetic correlation estimate, albeit highly uncertain (−0.8 ±1.5). A hypothesis is that BROOD and RESERVES reflect the proportion of a frame either filled with honey or brood. Consequentially, there is an antagonism between the two traits, as frames completely filled with honey cannot be filled with capped brood and vice-versa.
In addition, for the moderately negative correlation (phenotypic and genetic) between RESERVES in early summer and HONEY, two hypotheses can be made. First, both traits might be negatively correlated indirectly because of the negative correlation between RESERVES and BROOD and the positive one between BROOD and HONEY. Second, when colonies produce a fixed amount of honey and store more honey in the brood chamber, then they must store less of it in the super, lowering the honey yield. A second hypothesis, however, is that variation in total honey production could create a positive correlation between HONEY and RESERVES. This positive correlation would be due to colonies having more honey in both the super and the brood chamber when producing more in total, assuming that fixed proportions are allocated to the brood chamber and the super (van Noordwijk and de Jong 1986). Furthermore, colonies that store more honey in the brood chamber could have an advantage by suffering less from honey flow shortages, in turn benefitting honey production during strong honey flows. Such events were observed by the beekeepers and motivated them to integrate the RESERVES trait to their breeding objective. However, this advantage during honey flow shortages might have been partially suppressed by the beekeeper’s managing techniques, who nourish apiaries in emergency when needed, thus partially masking the expected effect of colonies with good RESERVES values on colony development and therefore on yield.
Lastly, HYG and VARROA had weak negative phenotypic (around −0.1) and genetic correlations, such that, to a small extent, selection for HYG may have an adverse effect on V. destructor load as also previously reported (Hoppe et al. 2020).
5 Conclusion
We reported results of a genetic analysis of a small honeybee population from an emerging breeding plan targeting the improvement of classical traits and traits related to resilience. In this initial phase, records were only taken on colonies of open-mated queens produced by controlled mated dams. The open-mated queens were only used as potential drone-producing queens. This created a structure in the data in which the queen effect on colony performance cannot be separated from the worker effect. For this reason, we used the Queen Model. Heritability estimates for production traits and traits related to resilience suggest that selection will be effective, although all our estimates had large standard errors due to the small dataset size. A genetic antagonism between the trait meTarpyasuring feed reserves and honey yield, however, might limit simultaneous improvement on both traits.
Data availability
The datasets generated or analyzed during the current study are available from the corresponding author upon reasonable request.
Code availability
The R code to obtain the inverse of honeybee-specific relationship matrices is available at the AINV-honeybees GitHub repository: https://github.com/Tristan-Kistler/AINV-honeybees/; and archived at: https://doi.org/10.5281/zenodo.7951334. The version used in this study is v19.
References
Adam B (1987) Beekeeping at Buckfast Abbey, 4th edn. Northern Bee Books, St, Mary’s Abbey, Buckfast, Devon, England
Andonov S, Costa C, Uzunov A, Bergomi P, Lourenco D, Misztal I (2019) Modeling honey yield, defensive and swarming behaviors of Italian honey bees (Apis mellifera ligustica) using linear-threshold approaches. BMC Genet 20(1):78. https://doi.org/10.1186/s12863-019-0776-2
Barrett T, Dowle M, Srinivasan A, Gorecki J, Chirico M, Hocking T (2024). data.table: Extension of 'data.frame'. R package version 1.15.99. https://Rdatatable.gitlab.io/data.table. https://github.com/Rdatatable/data.table. https://r-datatable.com
Basso B, Kistler T, Phocas F (2024) Genetic parameters, trends, and inbreeding in a honeybee breeding program for royal jelly production and behavioral traits. Apidologie 55(1):11. https://doi.org/10.1007/s13592-023-01055-3
Baudry E, Solignac M, Garnery L, Gries M, Cornuet J, Koeniger N (1998) Relatedness among honeybees (Apis mellifera) of a drone congregation. Proc R Soc Lond B 265(1409):2009–2014. https://doi.org/10.1098/rspb.1998.0533
Bienefeld K, Hoppe A (2024) BeeBreed.eu—the web solution of honeybee breeding. BeeBreed.eu. www.beebreed.eu. Accessed 5 Apr 2024
Bienefeld K, Reinhardt F, Pirchner F (1989) Inbreeding effects of queen and workers on colony traits in the honey bee. Apidologie 20(5):439–450. https://doi.org/10.1051/apido:19890509
Brascamp EW, Bijma P (2014) Methods to estimate breeding values in honey bees. Genet Sel Evol 46(1):53. https://doi.org/10.1186/s12711-014-0053-9
Brascamp EW, Bijma P (2019) Software to facilitate estimation of genetic parameters and breeding values for honey bees. Proceedings of the 46th Apimondia 8–12 September 2019; Montréal, Canada. https://edepot.wur.nl/546515. Last Access 01 Sept 2023
Brascamp EW, Willam A, Boigenzahn C, Bijma P, Veerkamp RF (2016) Heritabilities and genetic correlations for honey yield, gentleness, calmness and swarming behaviour in Austrian honey bees. Apidologie 47(6):739–748. https://doi.org/10.1007/s13592-016-0427-9
Brascamp EW, Willam A, Boigenzahn C, Bijma P, Veerkamp RF (2018) Correction to: Heritabilities and genetic correlations for honey yield, gentleness, calmness and swarming behaviour in Austrian honey bees. Apidologie 49:462–463. https://doi.org/10.1007/s13592-018-0573-3
Büchler R, Andonov S, Bienefeld K, Costa C, Hatjina F, Kezic N, Kryger P, Spivak M, Uzunov A, Wilde J (2013) Standard methods for rearing and selection of Apis mellifera queens. J Apic Res 52(1):1–30. https://doi.org/10.3896/IBRA.1.52.1.07
Büchler R, Kovačić M, Buchegger M, Puškadija Z, Hoppe A, Brascamp EW (2020) Evaluation of traits for the selection of Apis mellifera for resistance against Varroa destructor. InSects 11(9):618. https://doi.org/10.3390/insects11090618
Chevalet C, Cornuet J-M (1982) Etude théorique sur la sélection du caractère «production de miel» chez l’abeille. I Modèle Génétique et Statistique Apidologie 13(1):39–65. https://doi.org/10.1051/apido:19820105
Cheverud JM (1988) A comparison of genetic and phenotypic correlations. Evolution 42(5):958–968. https://doi.org/10.1111/j.1558-5646.1988.tb02514.x
Colditz IG, Hine BC (2016) Resilience in farm animals: biology, management, breeding and implications for animal welfare. Anim Prod Sci 56(12):1961–1983. https://doi.org/10.1071/AN15297
Czekońska K, Łopuch S, Miścicki S, Bańkowski J, Szabla K (2023) Monitoring of hive weight changes in various landscapes. Apidologie 54(3):30. https://doi.org/10.1007/s13592-023-01012-0
Danka RG, Harris JW, Dodds GE (2016) Selection of VSH-derived “Pol-line” honey bees and evaluation of their Varroa-resistance characteristics. Apidologie 47(3):483–490. https://doi.org/10.1007/s13592-015-0413-7
Decourtye A, Mader E, Desneux N (2010) Landscape enhancement of floral resources for honey bees in agro-ecosystems. Apidologie 41(3):264–277. https://doi.org/10.1051/apido/2010024
Dietemann V, Nazzi F, Martin SJ, Anderson DL, Locke B, Delaplane KS, Wauquiez Q, Tannahill C, Frey E, Ziegelmann B, Rosenkranz P, Ellis JD (2013) Standard methods for Varroa research. J Apic Res 52(1):1–54. https://doi.org/10.3896/IBRA.1.52.1.09
Du M, Bernstein R, Hoppe A, Bienefeld K (2022) Influence of model selection and data structure on the estimation of genetic parameters in honeybee populations. G3 (Bethesda) 12(2):jkab450. https://doi.org/10.1093/g3journal/jkab450
Eynard SE, Sann C, Basso B, Guirao A-L, Le Conte Y, Servin B, Tison L, Vignal A, Mondet F (2020) Descriptive analysis of the Varroa non-reproduction trait in honey bee colonies and association with other traits related to Varroa resistance. InSects 11(8):492. https://doi.org/10.3390/insects11080492
Gohel D (2024) rvg: R Graphics Devices for 'Office' Vector Graphics Output. R package version 0.3.3. https://github.com/davidgohel/rvg
Gohel D, Moog S (2024) officer: Manipulation of Microsoft Word and PowerPoint Documents. R package version 0.6.6. https://CRAN.R-project.org/package=officer
Goulson D, Nicholls E, Botías C, Rotheray EL (2015) Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347(6229):1255957. https://doi.org/10.1126/science.1255957
Grosjean P, Ibanez F, Etienne M (2018) pastecs: package for analysis of space-time ecological series. R package version 1.4.2. https://CRAN.R-project.org/package=pastecs
Guichard M, Dietemann V, Neuditschko M, Dainat B (2020a) Advances and perspectives in selecting resistance traits against the parasitic mite Varroa destructor in honey bees. Genet Sel Evol 52(1):71. https://doi.org/10.1186/s12711-020-00591-1
Guichard M, Neuditschko M, Soland G, Fried P, Grandjean M, Gerster S, Dainat B, Bijma P, Brascamp EW (2020b) Estimates of genetic parameters for production, behaviour, and health traits in two Swiss honey bee populations. Apidologie 51(5):876–891. https://doi.org/10.1007/s13592-020-00768-z
Henderson CR (1973) Sire evaluation and genetic trends. J Anim Sci 1973(Symposium):10–41. https://doi.org/10.1093/ansci/1973.Symposium.10
Hernandez J, Maisonnasse A, Cousin M, Beri C, Le Quintrec C et al (2020) ColEval: honeybee COLony structure EVALuation for field surveys. InSects 11(1):41. https://doi.org/10.3390/insects11010041
Hoppe A, Du M, Bernstein R, Tiesler F-K, Kärcher M, Bienefeld K (2020) Substantial genetic progress in the international Apis mellifera carnica population since the Implementation of genetic evaluation. InSects 11(11):768. https://doi.org/10.3390/insects11110768
Houle D (1992) Comparing evolvability and variability of quantitative traits. Genetics 130(1):195–204. https://doi.org/10.1093/genetics/130.1.195
Kistler T, Brascamp EW, Basso B, Bijma P, Phocas F (2024) Uncertainty in the mating strategy of honeybees causes bias and unreliability in the estimates of genetic parameters. Genet Sel Evol 56(1). https://doi.org/10.1186/s12711-024-00898-3
Kretzschmar A, Maisonnasse A (2022) More worker capped brood and honey bees with less Varroa load are simple precursors of colony productivity at beekeepers’ disposal: an extensive longitudinal survey. InSects 13(5):472. https://doi.org/10.3390/insects13050472
Misztal I, Tsuruta S, Strabel T, Auvray B, Druet T, Lee DH (2002) BLUPF90 and related programs (BGF90). Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, pp 19−23
R Core Team (2021) R: A language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria
Rinderer TE, Harris JW, Hunt GJ, De Guzman LI (2010) Breeding for resistance to Varroa destructor in North America. Apidologie 41(3):409–424. https://doi.org/10.1051/apido/2010015
Roff DA (1995) The estimation of genetic correlations from phenotypic correlations: a test of Cheverud’s conjecture. Heredity 74(5):481–490. https://doi.org/10.1038/hdy.1995.68
Ruttner F (2013) Biogeography and taxonomy of honeybees. Springer Science & Business Media. ISBN number: 978-3-642-72649-1
Schlüns H, Moritz RFA, Neumann P, Kryger P, Koeniger G (2005) Multiple nuptial flights, sperm transfer and the evolution of extreme polyandry in honeybee queens. Anim Behav 70(1):125–131. https://doi.org/10.1016/j.anbehav.2004.11.005
Tarpy DR, Nielsen DI (2002) Sampling error, effective paternity, and estimating the genetic structure of honey bee colonies (Hymenoptera: Apidae). Ann Entomol Soc Am 95(4):513–528. https://doi.org/10.1603/0013-8746(2002)095[0513:SEEPAE]2.0.CO;2
Tarpy DR, vanEngelsdorp D, Pettis JS (2013) Genetic diversity affects colony survivorship in commercial honey bee colonies. Naturwissenschaften 100(8):723–728. https://doi.org/10.1007/s00114-013-1065-y
van Noordwijk AJ, de Jong G (1986) Acquisition and allocation of resources: their influence on variation in life history tactics. Am Nat 128(1):137–142. https://doi.org/10.1086/284547
Wei T, Simko V (2021) R package 'corrplot': Visualization of a correlation matrix. R package version 0.92. https://github.com/taiyun/corrplot
Wickham H, Averick M, Bryan J, Chang W, McGowan LD et al (2019) Welcome to the Tidyverse. J Open Source Softw 4(43):1686. https://doi.org/10.21105/joss.01686
Ždiniaková T, Loerchner C, De RO, Dimitrova T, Kaklamanos G et al (2023) EU Coordinated action to deter certain fraudulent practices in the honey sector. In: JRC Publications Repository. https://publications.jrc.ec.europa.eu/repository/handle/JRC130227. Accessed 6 Nov 2023
Acknowledgements
We thank all the beekeepers managing the hives and the breeding plan with the support of the ADAPI association and, in particular, its director, Pascal Jourdan, and staff members, in particular Robin Buisson, Alban Maisonnasse, and Emilie Tourlet, who helped phenotyping the colonies. We also thank INRAE staff Emilien Rottier for his help, primarily in phenotyping. We are also thankful to Naïs Rocher, Rodolphe Dupain, Louis Woerly, and Estelle Pignalet for their contributions in collecting phenotypic data through their internship projects and performing preliminary analysis on the pin-test records and feed reserves.
Funding
This work is supported by the CARNOT France Future Elevage (project BeeMuSe) and a public grant overseen by the French National Research Agency (ANR) as part of the « Investissements d’Avenir» program, through the “ADI 2021” project funded by the IDEX Paris-Saclay (ANR-11-IDEX-0003–02). The ADAPI breeding scheme is supported by the Sud-PACA Region, by FranceAgrimer, and by the departmental councils of Bouches-du-Rhône, Vaucluse, Alpes-de-Haute-Provence, and Alpes-Maritimes departments.
Author information
Authors and Affiliations
Contributions
TK participated in the bee phenotyping and the data curation, was in charge of the statistical analysis and the interpretation of results, and wrote the original draft. CK, CD, and BB participated in the bee phenotyping and data curation. CK, BB, FM, AV, and FP were involved in the conception of the study. PB, EWB, and FP supervised the work and were involved in the methodological choices, interpretation of the results, and revisions to the draft. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not required. The work involved data collected for farming purposes on an unregulated and very common insect species (Apis mellifera).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Manuscript editor: David Tarpy
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kistler, T., Kouchner, C., Brascamp, E.W. et al. Heritability and correlations for honey yield, handling ease, brood quantity, and traits related to resilience in a French honeybee population. Apidologie 55, 52 (2024). https://doi.org/10.1007/s13592-024-01088-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13592-024-01088-2