Abstract
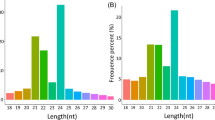
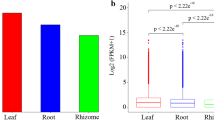
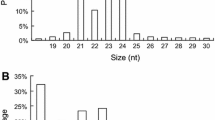
MicroRNAs (miRNAs) are a group of endogenous small non-coding RNAs with important roles in plant growth, development, and metabolic processes. Polygonatum cyrtonema Hua (P. cyrtonema) is an important Chinese traditional medicinal herb with broad pharmacological functions, and polysaccharides are the main biological substance accumulated in the P. cyrtonema rhizome. However, regulation of the process of polysaccharide biosynthesis in P. cyrtonema remains largely unknown.To elucidate the miRNAs and their targets involved in polysaccharide biosynthesis in P. cyrtonema, four small RNA libraries were constructed from flower, leaf, rhizome, and root tissues and sequenced. A total of 69 conserved and 5 novel miRNAs were identified, of which 6 miRNAs (miR156a-5p, miR156f-5p, miR395a-5, miR396a-3p, miR396g-3p, and miR397-5p-1) were down-regulated and 7 miRNAs (miR160, miR160h-1, miR160e-5p, miR319b-1, miR319-1, miR319c-5p-3, and miR319c-1) were up-regulated in rhizomes compared with flower, leaf, and root tissues. Bioinformatics analysis showed that the predicted targets of these miRNAs were mostly transcription factors and functional genes enriched in metabolic and secondary metabolite biosynthetic pathways, and 7 genes and their paired miRNAs were identified in carbohydrate metabolism. qRT-PCR expression analysis demonstrated that 6 miRNAs and their targets involved in carbohydrate metabolism were existed a negative correlation in. P. cyrtonema tissues. MiR396a-3p and one of its target genes, abfA, were possibly involved in polysaccharide biosynthesis pathway. This is the first report on the identification of conserved and novel miRNAs and their potential targets in P. cyrtonema, thus providing molecular evidence for the role of miRNAs in the regulation of polysaccharide biosynthesis in P. cyrtonema.







Similar content being viewed by others
Data availability
The sRNA-Seq data sets of four P. cyrtonema tissues have been deposited in NCBI Sequence Read Archive (SRA) database under the accession number SRP193031. The RNA-seq data sets of four P. cyrtonema tissues have been deposited in NCBI Sequence Read Archive (SRA) database under the accession number SRP193176.
Abbreviations
- miRNA:
-
MicroRNA
- qRT-PCR:
-
Quantitative real-time PCR
- DEM:
-
Differentially expressed miRNA
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- sRNA:
-
Small RNA
- TPM:
-
Transcripts per kilobase million
- SPL:
-
Squamosa promoter-binding protein-like
- ARF:
-
Auxin response factor
- MYB:
-
V-myb avian myeloblastosis viral oncogene homolog
- TCP:
-
Teosinte branched1/Cincinnata/proliferating cell factor
- abfA:
-
Alpha-L-arabinofuranosidase
- GalDH:
-
L-galactose dehydrogenase
- SORD:
-
L-iditol 2-dehydrogenase
- SDHA:
-
Succinate dehydrogenase
- PSPs:
-
Polygonatum polysaccharides
- GRF1:
-
Growth-regulating factor1
- SAMT:
-
Salicylic acid carboxyl methyltransferase
- GHs:
-
Glycosyl hydrolases
- NBS-LRR:
-
Nucleotide-binding site-leucine-rich repeat
References
Audic S, Claverie JM (1997) The significance of digital gene expression profiles. Genome Res 7(10):986–995
Bonar N, Liney M, Zhang R et al (2018) Potato miR828 is associated with purple tuber skin and flesh color. Front Plant Sci 9:1742
Chen L, Luan Y, Zhai J et al (2015) Sp-miR396a-5p acts as a stress-responsive genes regulator by conferring tolerance to abiotic stresses and susceptibility to Phytophthora nicotianae infection in transgenic tobacco. Plant Cell Rep 34(12):2013–2025
Chen L, Meng J, Zhai J et al (2017) MicroRNA396a-5p and -3p induce tomato disease susceptibility by suppressing target genes and upregulating salicylic acid. Plant Sci Int J Exp Plant Biol 265:177–187
Cui X, Wang S, Cao H et al (2018) A review: the bioactivities and pharmacological applications of polygonatum sibiricum polysaccharides. Molecules 23(5):1170
Damodharan S, Corem S, Gupta SK et al (2018) Tuning of SlARF10A dosage by sly-miR160a is critical for auxin-mediated compound leaf and flower development. Plant J Cell Mol Biol 96(4):855–868
Evers M, Huttner M, Dueck A et al (2015) miRA: adaptable novel miRNA identification in plants using small RNA sequencing data. BMC Bioinform 16:370
Fahlgren N, Carrington JC (2010) miRNA target prediction in plants. Methods Mol Biol 592:51–57
Fan W, Zhang S, Du H et al (2014) Genome-wide identification of different dormant Medicago sativa L. MicroRNAs in response to fall dormancy. PLoS ONE 9(12):e114612
Friedlander MR, Chen W, Adamidi C et al (2008) Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26(4):407–415
Gebelin V, Leclercq J et al (2013) The small RNA profile in latex from Hevea brasiliensis trees is affected by tapping panel dryness. Tree Physiol 33(10):1084–1098
Guan X, Pang M, Nah G et al (2014) miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun 5:3050
Hou J, Lu D, Mason AS et al (2019) Non-coding RNAs and transposable elements in plant genomes: emergence, regulatory mechanisms and roles in plant development and stress responses. Planta 250(1):23–40
Koyama T, Sato F, Ohme-Takagi M (2017) Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol 175(2):874–885
Langmead B, Trapnell C, Pop M et al (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25
Lee CH, Carroll BJ (2018) Evolution and diversification of small RNA Pathways in flowering plants. Plant Cell Physiol 59(11):2169–2187
Li XY, Lin EP, Huang HH et al (2018) Molecular Characterization of SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Gene Family in Betula luminifera. Front Plant Sci 9:608
Li W, Xu R, Yan X et al (2019) De novo leaf and root transcriptome analysis to explore biosynthetic pathway of Celangulin V in Celastrus angulatus maxim. BMC Genom 20(1):7
Li W, He Z, Zhang L et al (2017) miRNAs involved in the development and differentiation of fertile and sterile flowers in Viburnum macrocephalum f. keteleeri. BMC Genom 18(1):783
Liu N, Dong L, Deng X et al (2018) Genome-wide identification, molecular evolution, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon L. BMC Plant Biol 18(1):336
Luo Y, Zhang X, Luo Z et al (2015) Identification and characterization of microRNAs from Chinese pollination constant non-astringent persimmon using high-throughput sequencing. BMC Plant Biol 15:11
Megha S, Basu U, Joshi RK et al (2018) Physiological studies and genome-wide microRNA profiling of cold-stressed Brassica napus. Plant Physiol Biochem PPB 132:1–17 `
Natarajan B, Kalsi HS, Godbole P et al (2018) MiRNA160 is associated with local defense and systemic acquired resistance against Phytophthora infestans infection in potato. J Exp Bot 69(8):2023–2036
Qu D, Yan F, Meng R et al (2016) Identification of MicroRNAs and their targets associated with fruit-bagging and subsequent sunlight re-exposure in the “granny smith” apple exocarp using high-throughput sequencing. Front Plant Sci 7:27
Shen EM, Singh SK, Ghosh JS et al (2017) The miRNAome of Catharanthus roseus: identification, expression analysis, and potential roles of microRNAs in regulation of terpenoid indole alkaloid biosynthesis. Sci Rep 7:43027
Song S, Hao L, Zhao P et al (2019) Genome-wide identification, expression profiling and evolutionary analysis of auxin response factor gene family in potato (Solanum tuberosum Group Phureja). Sci Rep 9(1):1755
Sun Y, Niu X, Fan M (2017) Genome-wide identification of cucumber green mottle mosaic virus-responsive microRNAs in watermelon. Adv Virol 162(9):2591–2602
t Hoen PA, Ariyurek Y, Thygesen HH et al (2008) Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucl Acids Res 36(21):e141
Wang L, Du H, Wuyun TN (2016) Genome-wide identification of MicroRNAs and their targets in the leaves and fruits of eucommia ulmoides using high-throughput sequencing. Front Plant Sci 7:1632
Wang W, Shi T, Ni X et al (2018) The role of miR319a and its target gene TCP4 in the regulation of pistil development in Prunus mume. Genome 61(1):43–48
Wang S, Wang B, Hua W et al (2017) De novo assembly and analysis of polygonatum sibiricum transcriptome and identification of genes involved in polysaccharide biosynthesis. Int J Mol Sci 18(9):1950
Wang C, Peng D, Zhu J et al (2019) Transcriptome analysis of Polygonatum cyrtonema Hua: identification of genes involved in polysaccharide biosynthesis. Int J Mol Sci 15:65
Wei R, Qiu D, Wilson IW et al (2015) Identification of novel and conserved microRNAs in Panax notoginseng roots by high-throughput sequencing. BMC Genom 16:835
Wu J, Jiang Y, Liang Y et al (2019) Expression of the maize MYB transcription factor ZmMYB3R enhances drought and salt stress tolerance in transgenic plants. Plant Physiol Biochem PPB 137:179–188
Wu HJ, Ma YK, Chen T et al (2012) PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucl Acids Res 40(Web Server issue):W22-8
Yelithao K, Surayot U, Lee JH et al (2016) RAW264.7 cell activating glucomannans extracted from rhizome of polygonatum sibiricum. Prev Nutr Food Sci 21(3):245–254
Zeng RF, Zhou JJ, Liu SR et al (2019) Genome-wide identification and characterization of SQUAMOSA-promoter-binding protein (SBP) genes involved in the flowering development of citrus clementina. Biomolecules 9(2):66
Zhai R, Zhao Y, Wu M et al (2019) The MYB transcription factor PbMYB12b positively regulates flavonol biosynthesis in pear fruit. BMC Plant Biol 19(1):85
Zhang H, Cao Y, Chen L et al (2015) A polysaccharide from Polygonatum sibiricum attenuates amyloid-beta-induced neurotoxicity in PC12 cells. Carbohyd Polym 117:879–886
Zhao X, Li J (2015) Chemical constituents of the genus Polygonatum and their role in medicinal treatment. Nat Prod Commun 10(4):683–688
Zhao P, Zhao C, Li X et al (2018b) The genus Polygonatum: a review of ethnopharmacology, phytochemistry and pharmacology. J Ethnopharmacol 214:274–291
Zhao L, Zhang X, Qiu Z et al (2018a) De novo assembly and characterization of the xenocatantops brachycerus transcriptome. Int J Mol Sci 19(2):520
Zhu X, Li Q, Lu F et al (2015) Antiatherosclerotic potential of rhizoma polygonati polysaccharide in hyperlipidemia-induced atherosclerotic hamsters. Drug Res 65(9):479–483
Acknowledgements
We thank the Beijing Genomics Institute for assistance with experiments.
Funding
The financial support for the development and completion of this project was provided by startup funds through Natural Science Research Grant of Higher Education of Anhui Province (Grant Number KJ2019A0476, KJ2018ZD028) and Natural Science Foundation of Anhui Province of China (Grant Number 2008085MH268, 2108085MH315). The National Key Research and Development Program (Grant Number 2017YFC1701600) and the National project cultivation fund of Anhui University of Chinese Medicine (Grant Number 2020PY02) provided experimental support. Publication costs were funded by Outstanding Youth general Project of Anhui Institution of Higher Education (Grant Number gxyq2019030).
Author information
Authors and Affiliations
Contributions
Project design: J.W.W. Experiments and data analysis: K.L.M., S.X.Z., Z.L.Q., C.K.W., and Y.Y.S. Manuscript preparation: K.L.M. Preparation of plant materials: Q.S.Y. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, K., Zhang, S., Zhao, L. et al. Identification, expression analysis, and potential roles of microRNAs in the regulation of polysaccharide biosynthesis in Polygonatum cyrtonema Hua. J. Plant Biochem. Biotechnol. 31, 925–937 (2022). https://doi.org/10.1007/s13562-022-00772-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13562-022-00772-7