Abstract
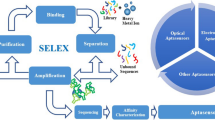
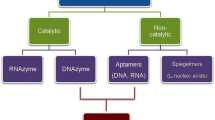
Aptamers are single-stranded oligonucleotides (DNA or RNA) capable of folding into well-defined structures and motifs that allow them bind to various target molecules with high specificity and affinity. The crucial ability of aptamers is interaction with a given target from small ions to molecular level that makes them useful in many applications for specific detection of various analytes. Nowadays aptamer is an effective tool not only for in vitro research in diagnostic methods, drug delivery and treatment therapy, but also for the development of nucleic-acid-based sensors in industrial waste management, pollution control and environmental toxicology. This mini review is focused on discussion and updating about the global researches in nucleic-acid-based sensors for environmental issues.
Similar content being viewed by others
References
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990).
Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510 (1990).
Song, K. M., Lee, S. & Ban, C. Aptamers and their biological applications. Sensors (Basel) 12, 612–631 (2012).
Zhou, J. & Rossi, J. J. Aptamer-targeted cell-specific RNA interference. Silence 1, 4 (2010).
Fang, X. & Tan, W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc. Chem. Res. 43, 48–57 (2010).
Ohuchi, S. Cell-SELEX Technology. Biores. Open Access 1, 265–272 (2012).
Meyer, S. et al. Development of an efficient targeted cell-SELEX procedure for DNA aptamer reagents. PLoS One 8, e71798 (2013).
Marimuthu, C., Tang, T. H., Tominaga, J., Tan, S. C. & Gopinath, S. C. Single-stranded DNA (ssDNA) production in DNA aptamer generation. Analyst 137, 1307–1315 (2012).
Dua, P., Kim, S. & Lee, D. K. Nucleic acid aptamers targeting cell-surface proteins. Methods 54, 215–225 (2011).
Kong, H. Y. & Byun, J. Nucleic Acid aptamers: new methods for selection, stabilization, and application in biomedical science. Biomol. Ther. (Seoul) 21, 423–434 (2013).
Mayer, G. et al. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 5, 1993–2004 (2010).
Lee, K.-A. et al. Aptamer-based sandwich assay and its clinical outlooks for detecting lipocalin-2 in hepatocellular carcinoma (HCC). Sci. Rep. 5, 10897 (2015).
Lee, S.-H. et al. Analytical bioconjugates, aptamers, enable specific quantitative detection of Listeria monocytogenes. Biosens. Bioelectron. 68, 272–280 (2015).
Wu, X., Chen, J., Wu, M. & Zhao, J. X. Aptamers: active targeting ligands for cancer diagnosis and therapy. Theranostics 5, 322–344 (2015).
Marolt, U., Cencic, A., Gorenjak, M. & Potrc, S. Generating Aptamers for Cancer Diagnosis and Therapy. Clin. Exp. Pharmacol. Physiol. 02, doi:10.4172/2161-1459.1000111 (2012).
Germer, K., Leonard, M. & Zhang, X. RNA aptamers and their therapeutic and diagnostic applications. Int. J. Biochem. Mol. Biol. 4, 27–40 (2013).
Simmons, S. C. et al. Anti-heparanase aptamers as potential diagnostic and therapeutic agents for oral cancer. PLoS One 9, e96846 (2014).
Huang, D.-B. et al. Crystal structure of NF-κB (p50) 2 complexed to a high-affinity RNA aptamer. Proc. Natl. Acad. Sci. USA 100, 9268–9273 (2003).
Farokhzad, O. C. et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc. Natl. Acad. Sci. USA 103, 6315–6320 (2006).
Tang, Z., Parekh, P., Turner, P., Moyer, R. W. & Tan, W. Generating aptamers for recognition of virus-infected cells. Clin. Chem. 55, 813–822 (2009).
Collett, J. R., Cho, E. J. & Ellington, A. D. Production and processing of aptamer microarrays. Methods 37, 4–15 (2005).
Bock, C. et al. Photoaptamer arrays applied to multiplexed proteomic analysis. Proteomics 4, 609–618 (2004).
Sekhon, S. S. et al. Advances in pathogen-associated molecules detection using Aptamer based biosensors. MCT 9, 311–317 (2013).
Yarus, M. How many catalytic RNAs? Ions and the Cheshire cat conjecture. FASEB J. 7, 31–39 (1993).
Pyle, A. M. Ribozymes: a distinct class of metalloenzymes. Science 261, 709–714 (1993).
Conrad, R. C., Giver, L., Tian, Y. & Ellington, A. D. in Methods in Enzymology Volume 591 (Academic Press, United States, 1996).
Higuchi, R. G. & Ochman, H. Production of single-stranded DNA templates by exonuclease digestion following the polymerase chain reaction. Nucleic Acids Res. 17, 5865 (1989).
Hultman, T., Stahl, S., Homes, E. & Uhlen, M. Direct solid phase sequencing of genomic and plasmid DNA using magnetic beads as solid support. Nucleic Acids Res. 17, 4937–4946 (1989).
Williams, K. P. & Bartel, D. P. PCR product with strands of unequal length. Nucleic Acids Res. 23, 4220–4221 (1995).
Breaker, R. R. Are engineered proteins getting competition from RNA? Curr. Opin. Biotechnol. 7, 442–448 (1996).
Tang, J. & Breaker, R. R. Rational design of allosteric ribozymes. Chem. Biol. 4, 453–459 (1997).
Penchovsky, R. Computational design of allosteric ribozymes as molecular biosensors. Biotechnol. Adv. 32, 1015–1027 (2014).
Breaker, R. R. Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotechnol. 13, 31–39 (2002).
Sook Bang, G. et al. Rational design of modular allosteric aptamer sensor for label-free protein detection. Biosens. Bioelectron. 39, 44–50 (2013).
Joyce, G. F. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 73, 791–836 (2004).
Breaker, R. R. & Joyce, G. F. A DNA enzyme with Mg2+-dependent RNA phosphoesterase activity. Chem. Biol. 2, 655–660 (1995).
O’Sullivan, C. K. Aptasensors -the future of biosensing? Anal. Bioanal. Chem. 372, 44–48 (2002).
Song, S., Wang, L., Li, J., Fan, C. & Zhao, J. Aptamer-based biosensors. Trends Anal. Chem. 27, 108–117 (2008).
Ostatná, V., Vaisocherová, H., Homola, J. & Hianik, T. Effect of the immobilisation of DNA aptamers on the detection of thrombin by means of surface plasmon resonance. Anal. Bioanal. Chem. 391, 1861–1869 (2008).
Hianik, T. & Wang, J. Electrochemical aptasensors - recent achievements and perspectives. Electroanalysis 21, 1223–1235 (2009).
Lu, Y. et al. New highly sensitive and selective catalytic DNA biosensors for metal ions. Biosens. Bioelectron. 18, 529–540 (2003).
Breaker, R. R. Catalytic DNA: in training and seeking employment. Nat. Biotechnol. 17, 422–423 (1999).
Chow, C. S. & Bogdan, F. M. A structural basis for RNA-ligand interactions. Chem. Rev. 97, 1489–1514 (1997).
Nowakowski, J., Shim, P. J., Prasad, G. S., Stout, C. D. & Joyce, G. F. Crystal structure of an 82-nucleotide RNA-DNA complex formed by the 10-23 DNA enzyme. Nat. Struct. Mol. Biol. 6, 151–156 (1999).
Ferguson, J. A., Steemers, F. J. & Walt, D. R. High-density fiber-optic DNA random microsphere array. Anal. Chem. 72, 5618–5624 (2000).
Taylor, L. C. & Walt, D. R. Application of high-density optical microwell arrays in a live-cell biosensing system. Anal. Biochem. 278, 132–142 (2000).
Li, J. & Lu, Y. A highly sensitive and selective catalytic DNA biosensor for lead ions. J. Am. Chem. Soc. 122, 10466–10467 (2000).
Klopman, G. in Chemical reactivity and reaction paths (Wiley, United States, 1974).
Da Silva, J. F. & Williams, R. J. P. in The biological chemistry of the elements: the inorganic chemistry of life (Oxford University Press, United Kingdom, 2001).
Neiboer, E & Richardson, D. H. S. The replacement of the nondescript term ‘heavy metals’ by biologically and chemically significant classification of metal. Environmental Pollution Series B, Chemical and Physical. 1, 3–26 (1980).
Needleman, H. Lead poisoning. Annu. Rev. Med. 55, 209–222 (2004).
Liu, J. & Lu, Y. A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J. Am. Chem. Soc. 125, 6642–6643 (2003).
Hoyle, I. & Handy, R. Dose-dependent inorganic mercury absorption by isolated perfused intestine of rainbow trout, Oncorhynchus mykiss, involves both amiloride-sensitive and energy-dependent pathways. Aquat. Toxicol. 72, 147–159 (2005).
Craft, E. S. et al. Depleted and natural uranium: chemistry and toxicological effects. J. Toxicol. Environ. Health Part B 7, 297–317 (2004).
Georgopoulos, P. et al. A framework and data sources for the assessment of human exposures to copper: The US Situation, http://ccl.rutgers.edu/ccl-files/reports/ICA/ICA2002_copper2.pdf (2002).
Xiang, Y., Tong, A. & Lu, Y. Abasic site-containing DNAzyme and aptamer for label-free fluorescent detection of Pb2+ and adenosine with high sensitivity, selectivity, and tunable dynamic range. J. Am. Chem. Soc. 131, 15352–15357 (2009).
Liu, C.-W., Huang, C.-C. & Chang, H.-T. Highly selective DNA-based sensor for lead (II) and mercury (II) ions. Anal. Chem. 81, 2383–2387 (2009).
Lee, J. S., Han, M. S. & Mirkin, C. A. Colorimetric Detection of Mercuric Ion (Hg2+) in Aqueous Media using DNA-Functionalized Gold Nanoparticles. Angew. Chem. 46, 4093–4096 (2007).
Wang, Y., Yang, F. & Yang, X. Colorimetric biosensing of mercury (II) ion using unmodified gold nanoparticle probes and thrombin-binding aptamer. Biosens. Bioelectron. 25, 1994–1998 (2010).
Lee, J. H., Wang, Z., Liu, J. & Lu, Y. Highly sensitive and selective colorimetric sensors for uranyl (UO2 2+): Development and comparison of labeled and label-free DNAzyme-gold nanoparticle systems. J. Am. Chem. Soc. 130, 14217–14226 (2008).
Yin, B.-C., Ye, B.-C., Tan, W., Wang, H. & Xie, C.-C. An allosteric dual-DNAzyme unimolecular probe for colorimetric detection of copper (II). J. Am. Chem. Soc. 131, 14624–14625 (2009).
Kim, M. et al. Arsenic removal from Vietnamese groundwater using the arsenic-binding DNA aptamer. Environ. Sci. Technol. 43, 9335–9340 (2009).
Wei, F. & Ho, C.-M. Aptamer-based electrochemical biosensor for Botulinum neurotoxin. Anal. Bioanal. Chem. 393, 1943–1948 (2009).
Zhou, L., Li, D.-J., Gai, L., Wang, J.-P. & Li, Y.-B. Electrochemical aptasensor for the detection of tetracycline with multi-walled carbon nanotubes amplification. Sens. Actuator B-Chem. 162, 201–208 (2012).
Zhang, J. et al. Label-free electrochemical detection of tetracycline by an aptamer nano-biosensor. Anal. Lett. 45, 986–992 (2012).
Chen, D., Yao, D., Xie, C. & Liu, D. Development of an aptasensor for electrochemical detection of tetracycline. Food Control 42, 109–115 (2014).
Cella, L. N. et al. Nano aptasensor for protective antigen toxin of anthrax. Anal. Chem. 82, 2042–2047 (2010).
Baeumner, A. J., Cohen, R. N., Miksic, V. & Min, J. RNA biosensor for the rapid detection of viable Escherichia coli in drinking water. Biosens. Bioelectron. 18, 405–413 (2003).
Sun, H., Choy, T., Zhu, D., Yam, W. & Fung, Y. Nano-silver-modified PQC/DNA biosensor for detecting E. coli in environmental water. Biosens. Bioelectron. 24, 1405–1410 (2009).
Cao, X. et al. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res. 37, 4621–4628 (2009).
Di Fusco, M., Tortolini, C., Frasconi, M. & Mazzei, F. Aptamer-based and DNAzyme-based biosensors for environmental monitoring. Int. J. Environ. Res. Public Health 5, 186–204 (2011).
Sett, A., Das, S. & Bora, U. Functional nucleic-acid-based sensors for environmental monitoring. Appl. Biochem. Biotechnol. 174, 1073–1091 (2014).
Xiao, Y., Lubin, A. A., Heeger, A. J. & Plaxco, K. W. Label-free electronic detection of thrombin in blood serum by using an aptamer-based sensor. Angew. Chem. 117, 5592–5595 (2005).
Radi, A.-E., Acero Sánchez, J. L., Baldrich, E. & O’Sullivan, C. K. Reagentless, reusable, ultrasensitive electrochemical molecular beacon aptasensor. J. Am. Chem. Soc. 128, 117–124 (2006).
Mir, M. & Katakis, I. Aptamers as elements of bioelectronic devices. Mol. Biosyst. 3, 620–622 (2007).
Bang, G. S., Cho, S. & Kim, B.-G. A novel electrochemical detection method for aptamer biosensors. Biosens. Bioelectron. 21, 863–870 (2005).
So, H.-M. et al. Single-walled carbon nanotube biosensors using aptamers as molecular recognition elements. J. Am. Chem. Soc. 127, 11906–11907 (2005).
Maehashi, K. et al. Label-free protein biosensor based on aptamer-modified carbon nanotube field-effect transistors. Anal. Chem. 79, 782–787 (2007).
He, P., Shen, L., Cao, Y. & Li, D. Ultrasensitive electrochemical detection of proteins by amplification of aptamer-nanoparticle bio bar codes. Anal. Chem. 79, 8024–8029 (2007).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Nguyen, PL., Sekhon, S.S., Ahn, JY. et al. Aptasensor for environmental monitoring. Toxicol. Environ. Health Sci. 9, 89–101 (2017). https://doi.org/10.1007/s13530-017-0308-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13530-017-0308-2