Abstract
Brook trout are a species of conservation concern in Southwestern Ontario, Canada, and effective monitoring of their populations is crucial for making informed management decisions. Electrofishing is a traditional, yet invasive, method that allows for fish abundance estimation. Environmental DNA (eDNA) is an emerging molecular tool that presents a non-invasive alternative to electrofishing. This study was a collaborative effort between researchers in academia, industry, and an NGO, with the following objectives: 1) compare eDNA detections with electrofishing when monitoring brook trout populations in a site of known occupancy, 2) compare existing eDNA collection methods, and 3) extend eDNA surveys to regions of unknown occupancy that could be of conservation concern (Hanlon Creek and Twelve Mile Creek, Ontario). First, eDNA sampling methods were validated with standard electrofishing. Water samples were filtered in tandem at each site using two commercially available eDNA samplers. The results suggest a significant difference in total eDNA capture and incidence of PCR inhibitors between the two autosamplers. Brook trout eDNA was detected at all locations in Hanlon Creek in September and November, as well as 5/6 sampling locations in Twelve Mile Creek. Brook trout signal in Hanlon Creek was stronger in November compared to September 2019, suggesting possible spawning activity. Brook trout eDNA was also detected in Twelve Mile Creek where brook trout were previously unreported. This study provides a technical validation for the use of eDNA in brook trout monitoring and illustrates the opportunity to use eDNA surveys in regulated settings to complement and improve conventional biomonitoring methods for the management of elusive species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A diverse freshwater biota is a vital indicator of watershed health, and understanding the trophic structure of the ecosystems in which aquatic organisms interact is crucial in the conservation of natural water bodies (Herman and Nejadhashemi 2015). Bony fish comprise a major trophic component in most freshwater ecosystems (Schindler and Scheuerell 2002). Salmonids are one family of bony fish that play an important role in creeks and streams as the link between terrestrial and aquatic food webs (Syrjänen et al. 2011). Some salmonids, such as brook trout (Salvelinus fontinalis), also act as bioindicators of freshwater ecosystem health, particularly in cold headwater streams (Johnson et al. 2016; Chadwick and McCormick 2017; Hermann et al. 2020). Consequently, they are of great importance to ecosystem function, creating a need for accurate and timely monitoring of their populations. Despite its success as an invasive species in Western North America (Larranga et al. 2018), the brook trout population is in decline in its native waters of Southeastern Ontario (Haxton et al. 2019). Being a cold-water species that relies on groundwater upwelling zones for spawning, the presence of brook trout has been viewed as an indicator of cool, clean, and undisturbed streams (Haxton et al. 2019).
It is thought that brook trout have not been able to adjust well to the thermal change in water quality that accompanies the large expansion of human built infrastructure in Southeastern Ontario (Wood 2017) and thus there is a need for effective population monitoring of the brook trout in this region.
Historically, bony fish have been monitored using direct capture methods, particularly via electrofishing, which uses an electrode to create electrical currents that stun fish (Schaeffer and Logan 2000). Fish are subsequently netted, placed into an aerated bucket, identified, counted, measured, and released. The invasive nature of the electrofishing method presents several drawbacks because the technique can cause considerable stress to organisms and the ecosystem, which can lead to unwanted mortality. Electrofishing may also be seasonally restricted due to the life histories of study organisms as they spawn, and potentially dangerous because prohibitive winter sampling conditions makes electrofishing in frozen environments undesirable (Schaeffer and Logan 2000; Thalinger et al. 2019). Furthermore, electrofishing surveys are time and labor intensive (Evans et al. 2017) and subject to regulatory permits (Jerde et al. 2013).
A novel, alternative approach to aquatic population monitoring is the use of environmental DNA (eDNA), which allows for the non-invasive detection of a species from an environmental sample (Lodge et al. 2012). Aquatic organisms release genetic material into their surrounding environment (eDNA) through dermal shedding and excretion, regurgitation, defecation, and other metabolic processes (Sassoubre et al. 2016). Additionally, eDNA can be accumulated following the release of gametes during spawning (Bracken et al. 2018). An eDNA signal can be retrieved from the water system via micro-filtration of water samples collected directly from the environment. The collection effort required for eDNA sampling is typically more efficient when compared to conventional techniques such as electrofishing (Evans et al. 2017). Additionally, eDNA can be used when target species are in life stages that may be sensitive to invasive monitoring methods (Thalinger et al. 2019). Collection and analysis of eDNA have aided global conservation efforts, particularly during monitoring of aquatic species at risk because it is typically more sensitive than conventional methods (Smart et al. 2015), making it particularly useful when monitoring species that are difficult to observe in their natural habitat (Beng and Corlett 2020). Furthermore, eDNA surveys can cover a broad geographic scale and access remote locations otherwise inaccessible with conventional surveys (Tucker et al. 2016).
The advantages and applications of eDNA technology are extensive. However, eDNA surveys have not yet been widely accepted by policy makers (i.e., government authorities) and the regulated community (i.e., industry, non-profits, private companies). This can be attributed to several limitations associated with targeted eDNA detection (Thomsen and Willerslev 2015; Beng and Corlett 2020), including the challenges posed by eDNA for directly estimating biomass and abundance of fish (Takahara et al. 2012; Pilliod et al 2013; Carraro et al. 2018; Bradley et al. 2022), and that eDNA can also be present in an ecosystem where the organism itself may not reside because of water movement carrying DNA from the organism of interest (Jane et al. 2014), or even from waste from other animals containing the DNA of interest (Jerde et al. 2013). Additionally, there is hesitancy to adopt targeted eDNA detection as a monitoring tool due to the perception of eDNA as an emerging field, and also because of the lack of standardized methods and reporting requirements (Evans et al. 2017; Jerde 2019; Loeza-Quintana et al. 2020; Petruniak et al. 2021). Before the regulated community and applied ecologists can redirect resources and budgets towards eDNA techniques, they need to be confident that eDNA results will be accepted by policy makers as standalones or complementary best practices (Petruniak et al. 2021). Researchers and applied scientists continue to identify opportunities, value, and limits of eDNA to inform and advance the application of this technology in a regulatory framework. Studies comparing eDNA surveys with conventional monitoring methods, recognizing challenges, and advancing eDNA technology are essential for standard setting. More importantly, collaborative efforts among researchers, applied ecologists, and the regulated community are critical for the acceptance and implementation of eDNA in regulated settings (Loeza-Quintana et al. 2020; Petruniak et al. 2021). In order to advance eDNA research, we present a collaborative study approaching three main elements: efficiency of eDNA for targeted salmonid detection, new technology, and application to biodiversity conservation.
The development and use of species-specific quantitative polymerase chain reaction (qPCR) assays for detecting eDNA in the context of environmental monitoring requires methodological validation (Thalinger et al. 2021). Many studies have applied qPCR to species-specific aquatic environmental monitoring (Berger and Aubin-Horth 2018; Harper et al. 2018; Thomas et al. 2020); however, relatively few have validated these methods by pairing eDNA collection with physical measurements of fish abundance and biomass (Wilcox et al. 2016; Kamoroff and Goldberg 2018; Knudsen et al. 2019). For example, Wilcox et al. (2016) found that qPCR analysis of eDNA was more sensitive than electrofishing with respect to the detection of rare piscine populations. Additionally, an eDNA survey of brook trout paired with electrofishing in a continuous high alpine lake system found that eDNA concentration acted as a predictor of target species population size (Kamoroff and Goldberg 2018). Evans et al. (2017) compared brook trout eDNA detection with electrofishing and found that eDNA decreased sampling effort and cost 67% less compared to triple-pass electrofishing, though eDNA cost more than presence/absence electrofishing and did not provide information on population structure (Evans et al. 2017).
Despite the broad implementation of eDNA methods in environmental studies, there is still a need for the standardization of methods for sample collection and analysis (Ruppert et al. 2019; Helbing and Hobbs 2019; Loeza-Quintana et al. 2020). The first objective of this study was to compare the accuracy of eDNA detections with electrofishing for biomonitoring brook trout populations. Many methodological approaches have been developed for collecting and processing eDNA samples, including portable technology that allows processing eDNA samples on-site, though this has resulted in a lack of standardized methods for eDNA sampling. One commercially available eDNA sampler is the portable filtration system developed by Smith‐Root (Thomas et al. 2018; Smith-Root 2020) which has been widely used (Thomas et al. 2018, 2020; Skinner et al. 2020; Pope et al. 2020; Loeza-Quintana et al. 2021). To inform best practices surrounding eDNA collection, the second objective of this study compared two eDNA portable filtration systems: the Smith-Root eDNA sampler (Thomas et al. 2018; Smith-Root 2020), and the OSMOS eDNA sampler from Halltech Aquatic (Halltech 2019). Both portable filtration systems are commercially available for use in eDNA monitoring, and both involve micro-filtration of water derived from environmental samples. The major difference between these sampling backpacks lies in the water uptake mechanism; the Smith-Root eDNA Sampler pulls water through a plastic tube (2.54 cm diameter), while the Halltech pulls water through the sides of a metal filter cartridge (Fig. 1). The Smith-Root eDNA sampler filter cartridges are disposable and made of plastic, featuring a pre-sterilized eDNA filter (Thomas et al. 2018, 2019) (Fig. 1). In contrast, the Halltech filter cartridges are made of aluminum and are reusable with many different types of filters, but these cartridges must be sterilized prior to use to avoid potential cross-contamination and the filters must be purchased separately (Halltech 2019) (Fig. 1). Use of the Smith-Root eDNA sampler has been reported in the previous scientific literature (Thomas et al. 2018, 2019, 2020; Gasparini et al. 2020; Pope et al. 2020; Skinner et al. 2020) while the use of the Halltech, to the authors’ best knowledge, has not.
Diagram of Smith-Root eDNA sampler (left) and Halltech (right) filter cartridges. Flow inlet and outlet are labelled along with 10 cm bar for scale. The Smith-Root eDNA sampler filter cartridge is comprised of a pre-sterilized filter and plastic cartridge attached to a 2.54-cm diameter tube which draws water vertically to pass through the filter housing. The Halltech filter cartridge requires insertion of a membrane prior to use and is made of aluminum with slots along the bottom edge that draw water in horizontally
This research was conducted in collaboration with Trout Unlimited Canada (TUC), a non-governmental organization (NGO) with the aim of conserving, protecting, and restoring Canadian freshwaters (Trout Unlimited Canada 2021). Industry partners also collaborated, namely the environmental consulting groups Natural Resource Solutions Inc. (NRSI) and SLR Consulting. All of the collaborators on this study were interested in monitoring brook trout (Salvelinus fontinalis) populations in the region. Brook trout are a native cold-water dwelling species that resides in southwestern Ontario streams and spawns in late autumn (Chadwick and McCormick 2017). Brook trout are of conservation concern in Ontario due to compounding habitat deterioration and developmental pressures, especially in the areas targeted in this study (Fig. 2) (Stephenson et al. 2009; Brenton 2010; Augustino et al. 2012). Effective and accurate monitoring of brook trout populations is integral to the conservation efforts of the species in these regions. Therefore, the third objective of our research was to employ eDNA surveys to aid monitoring efforts in regions of conservation concern (Hanlon Creek and Twelve Mile Creek) in Ontario, Canada (Fig. 2).
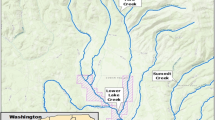
a Map depicting the five locations sampled along Hanlon Creek in both September and November of 2019. Map was generated using the Atlas of Canada—Toporama online tool (Ministry of Natural Resources Canada 2021). Each site is labeled with a number encapsulated in a pale blue circle, creek footprint is outlined in dark blue, and flow direction is indicated with dark blue arrows. Contour lines showing elevation are provided in brown, municipal development in red, and vegetation in green. For metadata associated with each sampling location (ex. latitude, longitude, and stream order), see Supplementary Material I. b: Map depicting the 6 locations sampled along the 12-Mile Creek watershed in November 2019. Map was generated using the Atlas of Canada—Toporama online tool (Ministry of Natural Resources Canada 2021). Each site is labeled with a number encapsulated in a pale blue circle, creek footprint is outlined in dark blue, and flow direction is indicated with dark blue arrows. Surrounding water bodies are indicated in cyan. Contour lines showing elevation are brown, and vegetation in green. For metadata associated with each sampling location (ex. latitude, longitude, and stream order), see Supplementary Material I
This study illustrates the opportunity to use eDNA surveys to complement and improve conventional methods of monitoring for the management of species at risk and/or invasive species. Our work included collaboration with industry partners, NGOs, scholars, and researchers, which represents an important step toward the acceptance and implementation of eDNA surveys in regulated settings. The interdisciplinary collaboration between diverse stakeholders and the novel validation of eDNA detection with electrofishing presented in this work are crucial steps towards integrating targeted eDNA surveys into conservation and management practices.
Methods
Field sampling methods
Sampling took place in two locales: 1) Preservation Park at Hanlon Creek in Guelph, Ontario on September 14th and November 4th, 2019; and 2) Twelve Mile Creek in the Niagara region of Ontario on November 23rd, 2019 (Fig. 2). The largest straight distance between sites at Hanlon Creek was about 1.3 km (between sites 1 and 5), while the largest straight distance between sites at Twelve Mile Creek was 6.2 km (between sites 1 and 5). These sampling locations were chosen due to the interest of TUC in these watersheds and to aid in the conservation efforts of brook trout in areas subject to developmental pressures and habitat deterioration. Sites were ranked as “brook trout expected” or “brook trout not expected” according to the Environmental Implementation Report on Hanlon Creek Business Park conducted by Natural Resources Solutions Inc. (Supplementary Material III, Table 1a-1b) (Stephenson et al. 2009; Brenton 2010). Five sites on Hanlon Creek were selected, each consisting of a 40-m transect (based on site accessibility) that had historically been electrofished by TUC. Six sites were chosen along Twelve Mile Creek which were also of interest to TUC. The Twelve Mile Creek sites were not assigned 40 m transects, as electrofishing did not occur there; only water filtration was required for eDNA sampling. The selection of these sites was also influenced by their accessibility, as many branches of this creek cross private property or were otherwise inaccessible.
The September sampling event in Hanlon Creek was the only event that included both electrofishing and eDNA sampling. In this event, eDNA sampling occurred prior to electrofishing to avoid cross-contamination or disturbance of brook trout. The electrofishing unit implemented in this study was the HallTech Aquatic HT2000 electrofishing backpack, with pulse frequency between 40 and 60 Hz. The electrofishing unit had one operator while two people (netters) netted emergent fish on either side. Electrofishing began downstream, moving upstream over the 40-m transect of each site using a single-pass method. Once fish were spotted, they were captured by netters and placed into an aerated bucket filled with local stream water. Once electrofishing was completed at each site, fish were identified and promptly placed back into the stream. Mortality due to electrofishing was not observed in any fish at any site during this process. This procedure was repeated at each site. Electrofishing could not be completed during either of the November sampling events on Hanlon Creek and Twelve Mile Creek because the brook trout spawning season was underway in the region. Spawning is a sensitive life cycle stage that could be negatively affected by the stress caused by electrofishing (Schaeffer and Logan 2000; Thalinger et al. 2019).
All water samples were collected from areas that had visibly free-flowing, clear water to avoid excessive sediment uptake and reduce filter clogging. Each sampling event was conducted within 12 h to ensure data from each sampling site was collected in relatively consistent conditions. Both eDNA portable filtration systems were sterilized with 5% bleach and rinsed with deionized water prior to use. To avoid potential cross-contamination of samples, Halltech filter cartridges were sterilized with 5% bleach and Eliminase, then sealed in clean plastic bags; pre-sterilized filter cartridges for the Smith-Root eDNA sampler were opened in the field immediately prior to eDNA collection.
Both eDNA samplers (Smith-Root and Halltech) were positioned in parallel along the stream bank at each site. One-liter water samples were filtered through 5-µm mixed cellulose ester (MCE) filters. The 5-µm filter pore size was chosen over the 1-µm alternative pore size because the turbidity of the creeks made filter clogging issues likely, so a larger pore size was deemed more suitable for this application. Using both Smith-Root and Halltech eDNA samplers, two samples were taken with each filtration system at each site. At site five in Hanlon Creek in September, an additional sample was filtered on the Smith-Root eDNA sampler due to a filter tearing. Field negative controls consisting of 1 L of sterilized tap water were filtered and processed using both the Smith-Root eDNA sampler and Halltech following the same protocol described above for field sample collection.
Metadata such as water and air temperature (°C), dissolved oxygen (DO) concentration (ppm), salinity (ppt), conductivity (uS□cm−1), flow velocity (m□s−1), and physical characteristics of each site were recorded (Supplementary Material I). To avoid potential cross-contamination of the water samples, all metadata parameters (temperature, DO, etc.) were measured after eDNA collection.
Field eDNA extractions
Filters were individually placed into 5-mL plastic vials containing lysis buffer solution and a steel ball bearing and shaken by hand for 60 s. DNA extractions were completed using the Biomeme M1 Sample Prep Cartridge Kit following the manufacturer's protocol (Biomeme 2021) using molecular methods described by Hermans et al. (2018). All extractions from the September Hanlon Creek and November at Niagara sampling events were completed in the field. Due to precipitation posing a contamination risk, extractions from the November Hanlon Creek sampling event were completed at the University of Guelph workspaces within 6 h of sample collection in a laboratory dedicated to eDNA processing that is isolated from high-concentration DNA. Samples of extracted DNA (in the cases of September and Niagara sampling events) and filters in lysis buffer (from the November sampling event) were then stored in a cooler on ice for transportation to the laboratory. Approximately 1 mL of the final elution derived from each Biomeme extraction was transferred into individually labeled plastic vials and then stored at − 20 °C.
eDNA detection using qPCR
Using similar methods described by Balasingham et al. (2018), Taqman qPCR analysis was conducted in the Hanner Laboratory at the University of Guelph using a proprietary species-specific brook trout TripleLock™ qPCR assay provided by Precision Biomonitoring Inc (Precision Biomonitoring 2019). The brook trout TripleLock™ qPCR assay targets a 90-bp region of the mitochondrial genome. All qPCR reactions were performed using the MIC (Bio Molecular Systems) thermocycler with a final reaction volume of 20 μL, including 5 μL of total DNA extracted from each environmental sample and following the manufacturer’s protocol (brook trout TripleLock™ qPCR assay; Precision Biomonitoring Inc). All qPCR reactions used LyoDNA™2.0 + IPC Master Mix (Biomeme 2021), which includes an internal positive control (IPC) to test for inhibition. The successful amplification and quantification of the IPC in an environmental sample suggests the lack of inhibitors in the sample. The absence of amplification or delayed amplification (≥ 3 cycles compared to the amplification in no-template control (NTC) samples) of the IPC is indicative of qPCR inhibition (Hartman et al. 2005). Four biological replicates were taken at each site, with two samples from each autosampling method. Six technical replicates were run per each biological sample (Buxton et al. 2021). A positive gBlock™ control (Integrated DNA Technologies 2021) and a negative no-template control (NTC) were included in each qPCR run. The various stages of qPCR were set up in spatially separated rooms to prevent contamination of eDNA samples, with all eDNA sample processing occurring in a designated clean room. All stages of qPCR set-up were carried out in a laminar flow UV hood (AirClean 600 PCR workstation), using 15 min of UV-decontamination of supplies and tools before setting up each reaction. Following qPCR, the raw MIC files were exported and input to MDMAPR 2.0 (Yu et al. 2020) for visualization of brook trout detections.
Statistical methods
To obtain estimates of eDNA concentration in the original samples, an eight-point standard curve of serial dilutions ranging in concentration from 1.33 × 10−1 to 1.33 × 106 copies per µL was created using known concentrations of synthetic brook trout gBlock DNA (IDT). The standard curve was analyzed once using the same targeted assay and qPCR method implemented on all samples (Fig. 3). According with Klymus et al. 2019, the limit of detection (LOD) was defined as the lowest standard concentration that can be detected with a 95% level of confidence. The limit of quantification (LOQ) was defined as the lowest standard concentration that could be quantified with a coefficient of variation (CV) value below 35%. The LOD and LOQ were both identified to be 133 copies/µL from the standard curve. The relationship between eDNA concentration and cycle of quantification (Cq) values is displayed in Eq. 1.
Standard curve of the brook trout TripleLock™ qPCR assay (Precision Biomonitoring 2019) illustrating the linear dynamic range of the assay. Assay has an efficiency (Efficiency = [10(-1/slope)]-1) of 85.3% and R2 = 0.9935. The limit of detection (LOD) and limit of quantification (LOQ) were both identified to be 133 copies/µL. LOD was defined as the lowest standard concentration that can be detected with a 95% level of confidence. The LOQ is the lowest standard concentration that could be quantified with a coefficient of variation (CV) value below 35% (Klymus et al. 2019). This figure was created using MIC qPCR Analysis Software
Equation. 1: Regression equation used to calculate brook trout eDNA concentration as a function of cycle of quantification, derived from the qPCR standard curve.
All data analysis was conducted in R Studio (version 1.1.463). Using the formula derived from the standard curve (Eq. 1), eDNA concentration was calculated from the Cq value of each technical qPCR replicate. Cq values are inversely proportional to brook trout eDNA concentration within the fluorescent range of the assay (determined to fall between 25 and 39 cycles for this assay). Thus, any Cq value outside of this range and its corresponding eDNA concentration were replaced with a zero. Additionally, a binary (yes/no) column representing whether or not each qPCR replicate showed inhibition based on the red IPC Cq values was created using the parameters outlined above in order to test the variation in inhibition between autosampling methods.
Preliminary data exploration showed a non-normal distribution of eDNA concentration, which was confirmed with a Shapiro–Wilk test for normality (W = 0.48112, p < 0.05). Thus, non-parametric contrasts were used to address the objectives of this study. To assess whether brook trout eDNA concentration correlated with the abundance of brook trout caught via electrofishing (objective 1), a Spearman non-parametric correlation test was used. This test was selected because the number of brook trout caught is an ordinal variable, while the eDNA concentration at each site was a continuous non-normal variable.
To compare the concentration of eDNA obtained between the two autosamplers, mean brook trout eDNA concentration at each sampling site and time was calculated in order to create a linear regression. Because the data was non-normal (Supplementary Material II), the data was transformed with the natural logarithm, and 0.5 was added to all eDNA concentration values prior to the transformation to avoid taking the natural log of zero. Following the natural log transformation, the assumptions for linear regression were tested (Supplementary Material II), and upon meeting the assumptions, a linear regression was constructed incorporating the mean eDNA concentration with the Halltech as the dependent variable and the mean eDNA concentration with the Smith-Root as the independent variable. To determine if inhibition varied significantly between autosamplers, a chi-square test was used to compare the binary inhibition measure (yes/no) between autosampler types. Finally, to determine if brook trout were present in Hanlon and Twelve Mile Creek (objective 3), the mean eDNA concentration for each site during each sampling event was calculated and plotted in boxplots. The number of brook trout caught via electrofishing during the September sampling event was also plotted on its corresponding boxplot.
Results
The brook trout TripleLock™ qPCR assay (Precision Biomonitoring 2019) was successful at detecting DNA from brook trout from water samples at Hanlon Creek and Twelve Mile Creek. All positive controls amplified as expected in every run, demonstrating proper performance of the assay and qPCR reaction. None of the negative controls, including field and laboratory NTC controls, yielded a positive detection. The results from the IPC control (delayed amplification ≥ 3 cycles compared to the amplification in NTC samples) suggest that qPCR inhibitors were present in September in three sites at Hanlon Creek. However, qPCR inhibition was only detected in samples collected using the Smith-Root eDNA sampler (Supplementary Material III, Table 1a). The LOD and LOQ were both identified to be 133 copies/µL from the standard curve (Fig. 3).
Objective 1: Comparison of eDNA and electrofishing for brook trout monitoring
During the Hanlon Creek September electrofishing event, brook trout were netted and identified at all sites except site 1 (Fig. 4). Most brook trout were caught at site 5, with a total of 10 animals caught. The identity and quantity of all fishes caught via electrofishing are provided in Supplementary Material IV; the number of species netted at sites 1, 2, 3, 4, and 5 were 5, 4, 4, 6, and 5, respectively, for a total of 137 fish netted. Brook trout eDNA was detected at all sites (Fig. 4). The Spearman’s correlation test showed no significant correlation between brook trout eDNA concentration and number of brook trout caught (S = 312 627, p = 0.4888) (Fig. 4).
Comparison between the eDNA concentration obtained through technical qPCR replicates on the MIC thermocycler and the number of Brook Trout caught via electrofishing at all five sites on Hanlon Creek in September 2019. Boxplot represents median (bold line), interquartile ranges (grey box), and upper and lower range of data (whiskers) with outliers labelled with white points outlined in black; the boxplot corresponds with the axis labels on the left, indicating the brook trout eDNA concentration in copies/μL. The number of fish caught via electrofishing at each site is indicated with a red diamond, and these points correspond with the red axis on the right side of the plot. A Spearman’s correlation test found no significant correlation between brook trout eDNA concentration and number of brook trout caught (S = 312 627, p = 0.4888)
Objective 2: Comparison of Smith-Root eDNA sampler and Halltech eDNA sampler portable filtration systems
Brook trout eDNA was detected using both the Smith-Root and Halltech eDNA samplers. To compare the DNA capture of both portable filtration systems, mean brook trout eDNA detected with each sampling apparatus at each site and time was calculated, then compared between autosamplers through a linear regression using the log-transformed values obtained from the Smith-Root as the independent variable, and the log-transformed values obtained from the Halltech as the dependent variable. The linear regression provided a y intercept (B0) of 1.0068, and a coefficient associated with the slope (B1) of 0.6284. Both values are significant (p < 0.05). This suggests a significant difference in total eDNA capture between the two autosamplers, and means that, for every 1% increase in the natural-log-transformed eDNA concentration obtained with the Smith-Root, the natural-log-transformed eDNA concentration obtained with the Halltech increases by 0.6272%. In other words, the results from this study suggest that the Smith-Root has higher eDNA capture efficiency than the Halltech.
To determine whether the autosamplers showed significant differences in the amount of qPCR inhibition, a Pearson’s Chi-square test with Yates’ continuity correction was used. This test compared the binary (yes/no) inhibition measure between the two sampling apparatuses and found that the inhibition varied significantly more than expected by chance between the two autosamplers (p < 0.05). Thus, the null hypothesis of this test, that the number of qPCR replicates with inhibition was the same between both autosamplers, was rejected. The results of this test suggest that eDNA samples collected with the Smith-Root autosampler showed significantly more inhibition than those collected with the Halltech.
Objective 3: Detection of brook trout eDNA in Hanlon Creek and Twelve Mile Creek via qPCR
The brook trout TripleLock™ qPCR assay (Precision Biomonitoring 2019) was used to detect brook trout eDNA in Hanlon Creek and Twelve Mile Creek (Fig. 5). Boxplots of eDNA concentration show that brook trout eDNA was present during all three sampling events (Figs. 6, 7, and 8). A summary of the results of targeted qPCR for brook trout in Hanlon Creek and Twelve Mile Creek is provided in Supplementary Material III, Table 1a (Hanlon Creek, September), Table 1b (Hanlon Creek, November) and Table 1c (Twelve Mile Creek, November). The qPCR results from all three sampling events were uploaded into the MDMAPR 2.0 platform (Yu et al. 2020) for visualization (Fig. 9). Additionally, a total of 15 adult brook trout were physically netted and observed in Hanlon Creek in September 2019 (Fig. 5).
Comparison of brook trout eDNA concentration detected via qPCR between two commercially available autosampling platforms, Smith-Root eDNA sampler, and Halltech eDNA sampler. Boxplot represents median (bold line), interquartile ranges (gray box), and upper and lower range of data (whiskers) with outliers labelled with white points outlined in black. A linear regression revealed a statistically significant relationship between the mean eDNA concentration (0.5 added to all values, then transformed with natural logarithm) between the Smith-Root (X) and the Halltech (Y) autosamplers at each sampling site and time (slope = 0.6284, p < 0.05)
Comparison of brook trout eDNA concentration detected in September at Hanlon Creek via qPCR between two commercially available sampling platforms, Smith-Root eDNA sampler and Halltech eDNA sampler, split by site. Boxplot represents median (bold line), interquartile ranges (gray box), and upper and lower range of data (whiskers) with outliers labelled with white points outlined in black. Samples filtered with the Smith-Root eDNA sampler are indicated in light grey, while samples filtered with the Halltech are indicated in dark gray. The qPCR LOD/LOQ is indicated with a horizontal red line at 133 copies/μL
Comparison of brook trout eDNA concentration detected in November at Hanlon Creek via qPCR between two commercially available sampling platforms, Smith-Root eDNA sampler and Halltech eDNA sampler, split by site. Boxplot represents median (bold line), interquartile ranges (gray box), and upper and lower range of data (whiskers) with outliers labelled with white points outlined in black. Samples filtered with the Smith-Root eDNA sampler are indicated in light gray, while samples filtered with the Halltech are indicated in dark gray. The qPCR LOD/LOQ is indicated with a horizontal red line at 133 copies/μL
Comparison of brook trout eDNA concentration detected in November at Twelve Mile Creek via qPCR between two commercially available sampling platforms, Smith-Root eDNA sampler, and Halltech eDNA sampler, split by site. Boxplot represents median (bold line), interquartile ranges (gray box), and upper and lower range of data (whiskers) with outliers labelled with white points outlined in black. Samples filtered with the Smith-Root eDNA sampler are indicated in light gray, while samples filtered with the Halltech are indicated in dark gray
Map depicting number of qPCR technical replicates derived from each of the creeks sampled (Hanlon Creek and Twelve Mile Creek) and their resulting signal intensity for brook trout. Map generated using MDMAPR 2.0 (Yu et al. 2020) with raw MIC files. Quantification cycle (Cq) strength is indicated by color, with both creeks exhibiting signal intensities in the moderate-strong range
Discussion
Hanlon Creek
Brook trout presence was confirmed in Hanlon Creek through electrofishing (Fig. 4) in September 2019. In addition, brook trout eDNA was detected at all sites (Figs. 6 and 7). These results indicate that the species is present in Hanlon Creek, and that biological material from this brook trout population was present in each sampled site. Most notably, electrofishing in September at site 5 resulted in the catch-and-release of ten individual brook trout, the highest of any sampling site. The eDNA concentration for this site indicated the presence of brook trout but was relatively consistent with concentrations for most of the other sites, which had lower numbers of brook trout captured (Fig. 4). This would suggest that although the eDNA sampling identified the presence of brook trout at site 5, the concentration of eDNA in the sample did not accurately represent their relative abundance at such a restricted spatial scale. Future effort is warranted to further delineate the relationship between target organism abundance and eDNA signal intensity.
During the September Hanlon Creek sampling event, only site 1 did not yield any brook trout from electrofishing. However, all sites yielded at-least some species of fishes (Supplementary Material IV), and the ability to capture species abundance and community composition is one advantage of electrofishing over targeted detection methods. This further reinforces the point that perhaps electrofishing and targeted eDNA detection methods are complementary, each with their own associated pros and cons. Furthermore, the only site (site 1) where brook trout were not captured by electrofishing had the closest proximity to a walking trail and a highway. This site was also observed to have little canopy cover. The lack of shade and runoff from the highway caused by these features may have adversely affected water temperature and quality (e.g., by increasing turbidity), which may result in the gravitation of brook trout upstream towards cooler and/or less disturbed areas of the creek (Chadwick et al. 2015). A positive relationship between road density and water temperature has been identified in Southwestern Ontario, as well as a negative relationship between road density and both diversity and richness of fish and benthic invertebrates (Wallace et al. 2013). Interestingly, site 1 had eDNA concentrations at similar levels to all other sites at this time point, suggesting brook trout eDNA is being transported downstream and may be a better predictor of habitat occupancy at larger spatial scales than electrofishing. Downstream transport of eDNA could be one reason that brook trout signal was detected at the downstream sites, though further research is needed to determine this.
During the November sampling event, brook trout eDNA concentration was much higher at site 5 relative to the September sampling event (Figs. 6 and 7). The variation between sites observed for this sampling event was driven by the high concentration of eDNA at site 5; since these sites were in the same location as those sampled in September, this variation was likely not due to spatial resolution between tributaries. Rather, this spike in eDNA concentration at site 5 in November may have been caused by preferential spawning activity in this tributary, which would imply that this region is of particular ecological importance. However, our study did not include visual spawning surveys; therefore, we cannot confirm that spawning activity was happening at the time of sampling. Future research on spawning activity in site 5 would allow for a better understanding of the cause of this eDNA spike.
Other environmental parameters that affect the longevity of eDNA could account for the observed variation in eDNA concentration between sites in November, including but not limited to canopy cover/light penetrance of the creek, water pH, dissolved oxygen (DO), or tannin concentration (Harrison et al. 2019). Some of these parameters (canopy cover, pH, DO) were measured in the current study, and there was no indication of correlation between those parameters and eDNA concentration. Tannin concentration, however, was not measured in this study; these molecules are associated with decaying leaves and have been shown to inhibit PCR in eDNA studies (Hunter et al. 2019). In this study, we used an IPC to evaluate the impact of inhibitors on our qPCR analysis. A sample was considered inhibited if a shift of ≥ 3 cycles in the Cq value of the IPC was observed compared with the Cq of the NTC IPC. Our results suggest that qPCR inhibition was present in samples collected during September at Hanlon Creek using the Smith-Root eDNA sampler. Notably, it was raining only when we sampled at sites 1 and 2, but not during our sampling at sites 3–5. The results from the IPC suggest PCR inhibition in samples from sites 1, 2, and 5. We observed sediment in the filters from the Smith-Root sampler (Supplementary Material V) and had difficulty extracting DNA due to clogging. It seems plausible that the inhibition observed in samples from sites 1, 2, and 5 may be due to the amount of sediment collected in the Smith-Root sampler filters, which may be related to the flow dynamics of the Smith-Root sampler cartridge itself (Fig. 1). Since inhibitors have been known to be carried in sediments (Buxton et al. 2017; Stoeckle et al. 2017; Turner et al. 2015), and the filters from the Smith Root sampler contained more sediment than their Halltech counterparts (Supplementary Material V), we believe this could explain the observed inhibition. Our findings highlight the significance of using an IPC to ensure that PCR inhibitors are not introducing the presence of false negative results. This consideration also highlights the importance of metadata collection during environmental DNA sampling (Nicholson et al. 2020).
Twelve Mile Creek
Brook trout eDNA was amplified in most sites sampled along Twelve Mile Creek, though at some of these sites, less than half of the technical qPCR replicates showed positive amplification (Supplementary Material III, Table 1c). The exception was at site 2, where no technical replicates indicated positive amplification. Despite not reaching the LOD/LOQ, there were still some technical replicates exhibiting positive amplification (Supplementary Material III, Table 1c); thus, it is reasonable to consider these as suspected positive detections. Considering the very low levels of DNA concentration that this assay is detecting, repetitive positive amplification of eDNA in multiple technical replicates of a biological replicate is grounds to suggest that there were brook trout present close to the sampling location and time. Moreover, it has been suggested that using a less stringent LOD (< 95% detection) would be more appropriate when working with low content eDNA samples (Sieber et al. 2020). If we used the definition proposed by Sieber et al. (2020), where the LOD is the lowest standard concentration with 50% detection rate, the LOD for brook trout in this study would be 13.3 copies/uL supporting our suspected positive detections. Furthermore, none of the negative controls resulted in positive amplification suggesting that cross-contamination was highly unlikely. Although we cannot discard the possibility that brook trout DNA might have been introduced to the sampled sites by other organisms (e.g., via waste), we consider that this potential source of target DNA might not be significant given the amount of positive technical replicates per site.
Our results indicate that brook trout are likely present in the Twelve Mile Creek system, and that they preferentially utilize the tributaries at site 1, 3, and 6 where higher eDNA concentrations were found. Brook trout DNA was also detected at sites 4 and 5, but with a lower detection rate. The only site where DNA from the species was not detected during our November sampling was at site 2. These findings could be validated in future studies by pairing environmental DNA monitoring with a catch-and-release based method, though this analysis was effective for the purposes of this study and yielded important management information of local conservation significance.
More variation in eDNA concentration between sites in Twelve Mile Creek compared to Hanlon Creek suggests a patchier distribution of brook trout DNA in Twelve Mile Creek; however, this may be due to the larger physical distance between sampling points (Fig. 2). Concentrated eDNA levels are more likely to occur when the habitat supports a high density of brook trout, as was observed in Hanlon Creek. Additionally, the sampled stretches of Twelve Mile Creek were much more developed than the sites chosen at Hanlon Creek, which indicates that habitat heterogeneity likely plays an important role in where brook trout can exist in this region (Studinski et al. 2012).
Technical implications
Environmental DNA monitoring using the PBI brook trout assay as a detection method for brook trout populations has been validated at a site of demonstrated occupancy (Hanlon Creek) with this study. It also validated the utility of two alternative eDNA sampling systems, as brook trout were successfully detected using qPCR-based eDNA technology in locations where the presence of the species was confirmed in tandem with electrofishing. Finally, the eDNA detection method was used to suggest the presence of brook trout in a region of unknown occupancy with conservation implications. Importantly, qPCR technology aided in the detection of brook trout eDNA in Twelve Mile Creek, an area where the species was previously undetected with electrofishing methods. The eDNA approach also allowed for monitoring of brook trout populations late into the season (November) with minimal habitat disturbance. This would not be possible when using conventional electrofishing techniques due to the sensitivity of populations to extraneous pressures during spawning season (Thalinger et al. 2019; Schaeffer and Logan 2000) when provincial regulations would prohibit the use of electrofishing. The ability to extend the monitoring season to any point in the year gives new opportunities and advantages to aquatic population monitoring (Thalinger et al. 2019; Bylemans et al. 2017). These findings should inform future conservation efforts in these regions.
Our linear regression suggested that the Smith-Root autosampler yielded higher average brook trout eDNA concentrations than the Halltech autosampler. However, the Smith-Root autosampler also yielded higher overall inhibition than the Halltech as per the results of the Chi-square test. It is known that fish eDNA can be found in higher concentrations in sediments compared with aqueous eDNA (Sakata et al. 2020). Therefore, the overall higher concentrations of brook trout eDNA retrieved with the Smith-Root eDNA autosampler, compared with the Halltech eDNA autosampler, could be due to the amount of species DNA present in the sediments observed in the filters from the Smith-Root autosampler. Furthermore, the flow dynamics of the Smith-Root sampler cartridge (Fig. 1) imply that the sample is collected closer to the bottom where the eDNA concentration is likely to be higher as the DNA settles into bottom substrates (Jo and Yamanaka 2022). Thus, according to the results, this study concluded that 1) the Smith-Root yielded higher detections in the watersheds sampled in this study and its use would be advantageous when sampling shallow waters, but 2) if sediment load/inhibitors are a concern, then the use of the Halltech is recommended. Since the Smith-Root eDNA sampler draws water straight up through the cartridge, while the Halltech draws water through the slots on the side, it is possible that inhibitory substances or sediments may have been selectively collected based on horizontal versus vertical hydrodynamic flow conditions within the filter cartridges. Future research should investigate the mechanisms underlying system-specific flow dynamics and their effect on eDNA recovery in aquatic systems. Additionally, the Halltech has heating capabilities and could be more effective for cold-weather sampling.
Aside from the advantages to eDNA monitoring mentioned above, there were some drawbacks to this technique in comparison with electrofishing. Considering eDNA monitoring is a relatively new technique, there are some gaps in the knowledge of characteristics of eDNA (Harrison et al. 2019). Due to an incomplete understanding of eDNA decay rates and transportation within aquatic systems, the spatial resolution of this type of monitoring is not as clearly defined as electrofishing. With electrofishing, precise abundance, spatial and temporal information is known about the animals in an area. With eDNA monitoring, it is difficult to know precisely where and when the animal has shed the detected eDNA, as well as the physical location of the organism itself. Hence, it may be a useful screening method when paired with electrofishing to establish a baseline. Future eDNA detection can then be contextualized using the verified baseline, and our initial detections can be strengthened by additional eDNA sampling events in future years.
An additional drawback of current on-site eDNA capturing techniques is the potential physical clogging of the filters that are used within the sampling apparatus to catch eDNA prior to DNA extraction. During the sampling, there were issues with clogged microfilters which increased the potential of retention of inhibitory compounds like tannins (Kumar et al. 2020). DNA extractions were also made more challenging by physical clogging of the DNA extraction kits. This is more likely to occur when there is an increased load of sediment in the water, which is a potential limitation to eDNA filtration efficiency. Further research should address this issue by considering implementing a pre-filter when working in aquatic systems with a high sediment load.
Studies that focus on technical validation are an important contribution to the field of applied ecology. Proof of concept experiments and validation of new eDNA methodological approaches are essential to provide support for the use of this tool in a regulatory framework. The present study found that targeted eDNA detection corroborates a conventional approach (electrofishing), providing support for the use of this technology in place of or alongside established monitoring methods.
Ecological implications
Brook trout are a valuable bioindicator of pristine headwaters due to their status as cold-water dwellers (Chadwick et al. 2015). The findings of this study support the need for conservation of cold-water habitat in Hanlon Creek in Guelph and Twelve Mile Creek in Niagara region due to the presence of brook trout DNA. Considering that brook trout were detected in both sampling locations, and at both time points in Hanlon Creek with increased eDNA signal suggestive of spawning, these locations have strong conservation value. Future studies should consider carrying out more robust sampling of these sites, including seasonally relevant time points and increased distances between sites to get a more complete profile of brook trout eDNA dynamics in these creek systems. One relevant avenue for future research lies in habitat occupancy modelling (Wilcox et al. 2018) and increased spatial sampling would support its implementation. Additionally, more intensive downstream sampling should be conducted to help inform whether the populations detected in this study are isolated from one another. This will allow for more accurate modelling and conservation of fish populations in a non-invasive manner.
Conclusion
Brook trout eDNA was detected throughout Hanlon Creek and at two main branches of Twelve Mile Creek. The use of the PBI kit for eDNA monitoring was validated in the field with electrofishing at a site of expected occupancy. Further, two eDNA samplers were compared (Smith-Root and Halltech), revealing the need to consider sampling depth, filter intake flow dynamics, and rate of filtration to minimize sediment loading of filters. Finally, brook trout eDNA presence was confirmed through qPCR at sites of unknown occupancy in Twelve Mile Creek. These findings have substantial implications with respect to aquatic conservation of brook trout in the city of Guelph and Niagara region, as well as the potential applications of eDNA to aquatic biodiversity monitoring of species at risk and invasive species. Here we have demonstrated the reliability of and confidence in three eDNA survey components. First, we demonstrated the success of a commercial assay to detect brook trout genetic material from environmental samples. Second, we demonstrated the successful performance of both eDNA samplers. Lastly, we validated the concurrence of the eDNA detection with fish capture using conventional methods. Furthermore, we showed the high sensitivity of eDNA surveys for species detection when conventional methods are not an accessible option (e.g., spawning season). This research serves as a pilot study that can be used to guide future research in aquatic biodiversity monitoring and will help establish confidence in eDNA surveys among applied ecologists, NGOs, and other potential users of eDNA technology.
Data availability
The qPCR data and environmental metadata associated with this report is available through the University of Guelph Borealis Data Repository at the following link: https://borealisdata.ca/dataset.xhtml?persistentId=doi:10.5683/SP3/7SB2NY.
Code availability
The R Code used for analysis of qPCR data and figure creation is available through the University of Guelph Borealis Data Repository at the following link: https://borealisdata.ca/file.xhtml?fileId=372495&version=1.0.
References
Augustino E, Baker J, Campbell J, Diamond J, Graszat A, Jamieson K, Lindblad D, McKinnon P, Metzger T, Stack M, Verkade G (2012) Twelve Mile Creek 2012 Watershed Report Card. Niagara Peninsula Conservation Authority: https://npca.ca/images/uploads/common/NPCA-2012-WatershedReportCard-Introduction.pdf. Accessed 31 January 2021
Balasingham KD, Walter RP, Mandrak NE, Heath DD (2018) Environmental DNA detection of rare and invasive fish species in two great lakes tributaries. Molec Ecol 27:112–127. https://doi.org/10.1111/mec.14395
Beng KC, Corlett RT (2020) Applications of environmental DNA (eDNA) in ecology and conservation: opportunities, challenges and prospects. Biodivers Conserv 29:1–33. https://doi.org/10.1007/s10531-020-01980-0
Berger CS, Aubin-Horth N (2018) An eDNA-qPCR assay to detect the presence of the parasite Schistocephalus solidus inside its threespine stickleback host. J Exp Biol 10:221. https://doi.org/10.1242/jeb.178137
Biomeme (2021) Biomeme Store. Biomeme Inc: https://shop.biomeme.com/. Accessed 31 January 2021
Bracken S, Rooney SM, Kelly-Quinn M, King JJ, Carlsson J (2018) Identifying spawning sites and other critical habitat in lotic systems using eDNA “snapshots”: a case study using the sea lamprey Petromyzon marinus. Ecol Evol 9:553–567. https://doi.org/10.1002/ece3.4777
Bradley DL, Morey KC, Borque DA, Fost B, Loeza-Quintana T, Hanner RH (2022) Environmental DNA detection and abundance estimates comparable to conventional methods for three freshwater larval species at a power plant discharge. Environ DNA 4:700–714. https://doi.org/10.1002/edn3.286
Brenton T (2010) Hanlon Creek Business Park Supplemental Environmental Implementation Report. Natural Resource Solutions Inc: https://guelph.ca/wp-content/uploads/HCBP_Supplemental-EIR-report.pdf. Accessed 31 Jan 2021
Buxton A, Groombridge J, Griffiths RA (2017) Is the detection of aquatic environmental DNA influenced by substrate type? PLoS ONE 12:e0183371. https://doi.org/10.1371/journal.pone.0183371
Buxton A, Matechou E, Griffin J, Diana A, Griffiths RA (2021) Optimising sampling and analysis protocols in environmental DNA studies. Sci Rep 11:11637. https://doi.org/10.1038/s41598-021-91166-7
Bylemans J, Furlan EM, Hardy CM, McGuffie P, Lintermans M, Gleeson D (2017) An environmental DNA-based method for monitoring spawning activity: a case study, using the endangered Macquarie perch (Macquaria australasica). Methods Ecol Evol 8:645–655. https://doi.org/10.1111/2041-210X.12709
Carraro L, Hartikainen H, Jokela J, Bertuzzo E, Rinaldo A (2018) Estimating species distribution and abundance in river networks using environmental DNA. Proc Natl Acad Sci USA 115:11724–11729. https://doi.org/10.1073/pnas.1813843115
Chadwick JG, Nislow KH, McCormick SD (2015) Thermal onset of cellular and endocrine stress responses corresponds to ecological limits in brook trout, an iconic cold-water fish. Conserv Physiol 3:1–12. https://doi.org/10.1093/conphys/cov017
Chadwick JG, McCormick SD (2017) Upper thermal limits of growth in brook trout and their relationship to stress physiology. J Exp Biol 220:3976–3987. https://doi.org/10.1242/jeb.161224
Evans NT, Shirey PD, Wieringa JG, Mahon AR, Lamberti GA (2017) Comparative cost and effort of fish distribution detection via environmental DNA analysis and electrofishing. Fisheries 42:90–99. https://doi.org/10.1080/03632415.2017.1276329
Gasparini L, Crookes S, Prosser RS, Hanner R (2020) Detection of freshwater mussels (Unionidae) using environmental DNA in riverine systems. Environ DNA 2:321–329. https://doi.org/10.1002/edn3.71
Halltech (2019) OSMOS eDNA sampler. Hall Tech Environmental and Aquatic Research Inc: http://halltech.ca/dir/products/osmos-edna-sampler/. Accessed 31 January 2021
Harper LR, Lawson Handley L, Hahn C, Boonham N, Rees HC, Gough KC, Lewis E, Adams IP, Brotherton P, Phillips S, Hänfling B (2018) Needle in a haystack? A comparison of eDNA metabarcoding and targeted qPCR for detection of the great crested newt (Triturus cristatus). Ecol Evol 8:6330–6341. https://doi.org/10.1002/ece3.4013
Harrison JB, Sunday JM, Rogers SM (2019) Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc Royal Soc b: Bio Sci 286:1–9. https://doi.org/10.1098/rspb.2019.1409
Hartman LJ, Coyne SR, Norwood D (2005) Development of a novel internal positive control for Taqman® based assays. Mol Cell Probes 19:51–59. https://doi.org/10.1016/j.mcp.2004.07.006
Haxton T, Ball H, Armstrong K (2019) Expert opinion on the status and stressors of brook trout, Salvelinus fontinalis, in Ontario. Fish Manag Ecol 27:111–122. https://doi.org/10.1111/fme.12376
Helbing CC, Hobbs J (2019) Environmental DNA standardization needs for fish and wildlife population assessments and monitoring. CSA Group: https://www.csagroup.org/wp-content/uploads/CSA-Group-Research-Environmental-DNA.pdf. Accessed 24 June 2021
Herman MR, Nejadhashemi AP (2015) A review of macroinvertebrate-and fish-based stream health indices. Ecohydrol Hydrobiol 15:53–67. https://doi.org/10.1016/j.ecohyd.2015.04.001
Hermann NT, Chaloner DT, Gerig BS, Lamberti GA (2020) Ecological consequences of Freat Lakes salmon subsidies for stream-resident brook and brown trout. Can J Fish Aquat Sci 77:1758–1771. https://doi.org/10.1139/cjfas-2020-0086
Hermans SM, Buckley HL, Lear G (2018) Optimal extraction methods for the simultaneous analysis of DNA from diverse organisms and sample types. Molec Ecol Resour 18:557–569. https://doi.org/10.1111/1755-0998.12762
Hunter ME, Ferrante JA, Meigs-Friend G, Ulmer A (2019) Improving eDNA yield and inhibitor reduction through increased water volumes and multi-filter isolation techniques. Sci Rep 9:5259. https://doi.org/10.1038/s41598-019-40977-w
Integrated DNA Technologies (2021) gBlocks Gene Fragments. Integrated DNA Technologies Inc: https://www.idtdna.com/pages/products/genes-and-gene-fragments/double-stranded-dna-fragments/gblocks-gene-fragments. Accessed 31 January 2021
Jane SF, Wilcox TM, McKelvey KS, Young MK, Schwartz MK, Lowe WH, Letcher BH, Whiteley AR (2014) Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Mol Ecol Resour 15:216–227. https://doi.org/10.1111/1755-0998.12285
Jerde CL, Chadderton WL, Mahon AR, Renshaw MA, Corush J, Budny ML, Mysorekar S, Lodge DM (2013) Detection of Asian carp DNA as part of a Great Lakes basin-wide surveillance program. Can J Fish Aquat Sci 70:522–526. https://doi.org/10.1139/cjfas-2012-0478
Jerde C (2019) Can we manage fisheries with the inherent uncertainty of eDNA? J Fish Biol 98:341–353. https://doi.org/10.1111/jfb.14218
Jo T, Yamanaka H (2022) Meta-analyses of environmental DNA downstream transport and deposition in relation to hydrogeography in riverine environments. Freshw Biol 00:1–11. https://doi.org/10.1111/fwb.13920
Johnson JH, Abbett R, Chalupnicki MA, Verdoliva F (2016) Seasonal habitat use of brook trout and juvenile steelhead in a Lake Ontario tributary. J Freshw Ecol 31:239–249. https://doi.org/10.1080/02705060.2015.1123655
Kamoroff C, Goldberg CS (2018) Environmental DNA quantification in a spatial and temporal context: a case study examining the removal of brook trout from a high alpine basin. Limnol 19:335–342. https://doi.org/10.1007/s10201-018-0551-5
Knudsen SW, Ebert RB, Hesselsøe M, Kuntke F, Hassingboe J, Mortensen PB, Thomsen PF, Sigsgaard EE, Hansen BK, Nielsen EE, Møller PR (2019) Species-specific detection and quantification of environmental DNA from marine fishes in the Baltic Sea. J Exp Mar Biol Ecol 510:31–45. https://doi.org/10.1016/j.jembe.2018.09.004
Klymus KE, Merkes CM, Allison MJ, Goldberg CS, Helbing CC, Hunter ME, Jackson CA, Lance RF, Mangan AM, Monroe EM, Piaggio AJ, Stokdyk JP, Wilson CP, Richter CA (2019) Reporting the limits of detection and quantification for environmental DNA assays. Environ DNA 2:271–282. https://doi.org/10.1002/edn3.29
Kumar G, Eble J, Gaither MR (2020) A practical guide to sample preservation and pre-PCR processing of aquatic environmental DNA. Molec Ecol Resour 20:29–39. https://doi.org/10.1111/1755-0998.13107
Larranga N, Wallerius ML, Guo H, Cucherousset J, Johnssen JI (2018) Invasive brook trout disrupt the diel activity and aggregation patterns of native brown trout. Can J Fish Aquat Sci 76:1052–1059. https://doi.org/10.1139/cjfas-2018-0110
Lodge DM, Turner CR, Jerde CL, Barnes MA, Chadderton L, Egan SP, Feder JL, Mahon AR, Pfrender ME (2012) Conservation in a cup of water: estimating biodiversity and population abundance from environmental DNA. Mol Ecol 21:2555–2558. https://doi.org/10.1111/j.1365-294X.2012.05600.x
Loeza-Quintana T, Abbott CL, Heath DD, Bernatchez L, Hanner RH (2020) Pathways to increase standards and competency of eDNA surveys (PISCeS) – advancing collaboration and standardization efforts in the field of eDNA. Environ DNA 2:255–260. https://doi.org/10.1002/edn3.112
Loeza-Quintana T, Crookes S, Li PY, Reid DP, Smith M, Hanner RH (2021) Environmental DNA detection of endangered and invasive species in Kejimkujik National Park and Historic Site. Genome 64:172–180. https://doi.org/10.1139/gen-2020-0042
Ministry of Natural Resources Canada (2021) The Atlas of Canada – Toporama. Government of Canada: https://atlas.gc.ca/toporama/en/index.html. Accessed 4 May 2021
Nicholson A, McIsaac D, MacDonald C, Gec P, Mason EB, Rein W, Wrobel J, de Boer M, Milian-Garcia Y, Hanner R (2020) An analysis of metadata reporting in freshwater environmental DNA calls for the development of best practice guidelines. Environ DNA 2:343–349. https://doi.org/10.1002/edn3.81
Pilliod DS, Goldberg CS, Arkle RS, Waits LP (2013) Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Can J Fish Aquat Sci 70:1123–1130. https://doi.org/10.1139/cjfas-2013-0047
Pope KL, Goldberg CS, Nelson NL, Cummings AK, Seaborn T, Piovia-Scott J (2020) Designing environmental DNA surveys in complex aquatic systems: backpack sampling for rare amphibians in Sierra Nevada meadows. Aquat Conserv 30:1975–1987. https://doi.org/10.1002/aqc.3444
Petruniak J, Bradley D, Kelly JM, Hanner RH (2021) Commentary: integrating environmental DNA into applied ecological practice. J Environ Sci Stud 11:6–11. https://doi.org/10.1007/s13412-020-00638-1
Precision Biomonitoring (2019) Precision Biomonitoring: Intelligent DNA Solutions. Precision Biomonitoring Inc: https://precisionbiomonitoring.com/. Accessed 31 January 2021
Ruppert KM, Kline RJ, Rahman MS (2019) Past, present and future perspectives of environmental DNA (eDNA) metabarcoding: a systematic review in methods, monitoring, and applications of global eDNA. Glob Ecol Conserv 17:1–49. https://doi.org/10.1016/j.gecco.2019.e00547
Sakata MK, Yamamoto S, Gotoh RO, Miya M, Yamanaka H, Minamoto T (2020) Sedimentary eDNA provides different information on timescale and fish species composition compared with aqueous DNA. Environ DNA 2:508–512. https://doi.org/10.1002/edn3.75
Sály P, Erős T, Takács P, Specziár A, Kiss I, Bíró P (2009) Assemblage level monitoring of stream fishes: the relative efficiency of single-pass vs. double-pass electrofishing. Fish Res 99:226–233. https://doi.org/10.1016/j.fishres.2009.06.010
Sassoubre LM, Yamahara KM, Gardner LD, Block BA, Boehm AB (2016) Quantification of environmental DNA (eDNA) shedding and decay rates for three marine fish. Environ Sci Technol 50:10456–10464. https://doi.org/10.1021/acs.est.6b03114
Schaeffer L, Logan D (2000) Guidelines for electrofishing water containing salmonids listed under the Endangered Species Act. National Oceanic and Atmospheric Administration Marine Fisheries Service: https://www.arlis.org/docs/vol1/230935595.pdf. Accessed 31 January 2021
Schindler DE, Scheuerell MD (2002) Habitat coupling in lake ecosystems. Oikos 98:177–189. https://doi.org/10.1034/j.1600-0706.2002.980201.x
Sieber N, Hartikainen H, Vorburger C (2020) Validation of an eDNA-based method for the detection of wildlife pathogens in water. Dis Aquat Org 141:171–184. https://doi.org/10.3354/dao03524
Skinner M, Murdoch M, Loeza-Quintana T, Crookes S, Hanner R (2020) A mesocosm comparison of laboratory-based and on-site eDNA solutions for detection and quantification of striped bass (Morone saxatilis) in marine ecosystems. Environ DNA 2:298–308. https://doi.org/10.1002/edn3.61
Smart AS, Tingley R, Weeks AR, van Rooyen AR, McCarthy MA (2015) Environmental DNA sampling is more sensitive than a traditional survey technique for detecting an aquatic invader. Ecol Appl 25:1944–1952. https://doi.org/10.1890/14-1751.1
Smith-Root (2020) eDNA sampler. Smith Root Inc: https://www.smith-root.com/edna/edna-sampler. Accessed 31 January 2021
Stephenson D, Coleman-Smith S, Green D, Badger N (2009) Hanlon Creek Business Park Environmental Implementation Report. Natural Resources Solution Inc: https://guelph.ca/wp-content/uploads/HCBP_EIR_Report.pdf. Accessed 31 January 2021
Stoeckle BC, Beggel S, Cerwenka AF, Motivans E, Kuehn R, Geist J (2017) A systematic approach to evaluate the influence of environmental conditions on eDNA detection success in aquatic ecosystems. PLoS One 12:e0189119. https://doi.org/10.1371/journal.pone.0189119
Studinski JM, Hartman KJ, Niles JM, Keyser P (2012) The effects of riparian forest disturbance on stream temperature, sedimentation, and morphology. Hydrobiologia 686:107–117. https://doi.org/10.1007/s10750-012-1002-7
Syrjänen J, Korsu K, Louhi P, Paavola R, Muotka T (2011) Stream salmonids as opportunistic foragers: the importance of terrestrial invertebrates along a stream-size gradient. Can J Fish Aquat Sci 68:2146–2156. https://doi.org/10.1139/f2011-118
Takahara T, Minamoto T, Yamanaka H, Doi H, Kawabata Z (2012) Estimation of fish biomass using environmental DNA. PLoS ONE 7:e35868. https://doi.org/10.1371/journal.pone.0035868
Teixeira-de Mello F, Kristensen EA, Meerhoff M, González-Bergonzoni I, Baattrup-Pedersen A, Iglesias C, Kristensen PB, Mazzeo N, Jeppesen E (2014) Monitoring fish communities in wadeable lowland streams: comparing the efficiency of electrofishing methods at contrasting fish assemblages. Environ Monit Assess 186:1665–1677. https://doi.org/10.1007/s10661-013-3484-9
Thalinger B, Wolf E, Traugott M, Wanzenböck J (2019) Monitoring spawning migrations of potamodromous fish species via eDNA. Sci Rep 9:1–11. https://doi.org/10.1038/s41598-019-51398-0
Thalinger B, Deiner K, Harper LR, Rees HC, Blackman RC, Sint D, Traugott M, Goldberg CS, Bruce K (2021) A validation scale to determine the readiness of environmental DNA assays for routine species monitoring. Environ DNA 00:1–14. https://doi.org/10.1002/edn3.189
Thomas AC, Howard J, Nguyen PL, Seimon TA, Goldberg CS (2018) eDNA Sampler: a fully integrated environmental DNA sampling system. Methods Ecol Evol 9:1379–1385. https://doi.org/10.1111/2041-210X.12994
Thomas AC, Nguyen PL, Howard J, Goldberg CS (2019) A self-preserving, partially biodegradable eDNA filter. Methods Ecol Evol 10:1136–1141. https://doi.org/10.1111/2041-210X.13212
Thomas AC, Tank S, Nguyen PL, Ponce J, Sinnesael M, Goldberg CS (2020) A system for rapid eDNA detection of aquatic invasive species. Environ DNA 2:261–270. https://doi.org/10.1002/edn3.25
Thomsen PF, Willerslev E (2015) Environmental DNA – an emerging tool in conservation for monitoring past and present biodiversity. Biol Conserv 183:4–18. https://doi.org/10.1016/j.biocon.2014.11.019
Trout Unlimited Canada (2021) Who we are. Trout Unlimited Canada: https://tucanada.org/trout-unlimited-canada-2/. Accessed 1 November 2021
Turner CR, Uy KL, Everhart RC (2015) Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biol Conserv 183:93–102. https://doi.org/10.1016/j.biocon.2014.11.017
Tucker AJ, Chadderton WL, Jerde CL, Renshaw MA, Uy K, Gantz C, Mahon AR, Bowen A, Strakosh T, Bossenbroek JM, Sieracki JL, Beletsky D, Bergner J, Lodge DM (2016) A sensitive environmental DNA (eDNA) assay leads to new insights on Ruffe (Gymnocephalus cernua) spread in North America. Biol Invasions 18:3205–3222. https://doi.org/10.1007/s10530-016-1209-z
Wallace AM, Croft-White MV, Moryk J (2013) Are Toronto’s streams sick? A look at the fish and benthic invertebrate communities in the Toronto region in relation to the urban stream syndrome. Environ Monit Assess 185:7857–7875. https://doi.org/10.1007/s10661-013-3140-4
Wilcox TM, McKelvey KS, Young MK, Sepulveda AJ, Shepard BB, Jane SF, Whiteley AR, Lowe WH, Schwartz MK (2016) Understanding environmental DNA detection probabilities: a case study using a stream-dwelling char Salvelinus fontinalis. Biol Conserv 194:209–216. https://doi.org/10.1016/j.biocon.2015.12.023
Wilcox TM, Young MK, McKelvey KS, Isaak DJ, Horan DL, Schwartz MK (2018) Fine-scale environmental DNA sampling reveals climate-mediated interactions between native fish species. Ecosphere 9:e02500. https://doi.org/10.1002/ecs2.2500
Wood J (2017) The conservation and management of brook trout in Ontario: past, present, and future. Latornell Conservation Symposium: http://www.latornell.ca/wp-content/uploads/files/presentations/2017/Latornell_2017_W3A_Jacquelyn_Wood.pdf. Accessed 25 June 2021
Yu J, Young RG, Deeth LE, Hanner RH (2020) Molecular detection mapping and analysis platform for R (MDMAPR) facilitating the standardization, analysis, visualization, and sharing of qPCR data and metadata. Peer J 8:e9974. https://doi.org/10.7717/peerj.9974
Acknowledgements
The authors of this manuscript would like to thank Giordana Scodellaro and Eric Heisey for their contributions to the field and laboratory work involved with this project. We also wish to thank Dr. Yoamel Milián-Garcia for his valuable feedback on early drafts of this manuscript. Thank you to Dr. Jarrett Phillips for his support with the statistical analysis of the data collected in this study. We would like to thank our collaborators at Trout Unlimited Canada, Halltech Aquatic, SLR Consulting, and Natural Resources Solutions Inc. for their valuable contributions to this manuscript. Finally, the authors wish to thank the anonymous reviewers and editorial staff at the Journal of Environmental Studies and Sciences for their valuable feedback during the preparation of this manuscript.
Funding
The authors sincerely thank David Stephenson at Natural Resources Solutions Inc. (NRSI) who provided significant funding for this research. Trout Unlimited Niagara chapter provided funding for PCR reagents facilitating the research in the Twelve-Mile Creek region.
NRSI, SLR Consulting Ltd, Trout Unlimited Canada, Niagara College, and HallTech provided important in-kind contributions including personnel, field assistance, and equipment.
The authors also wish to thank Shoeless Joe’s Sports Grill on Clair Road in Guelph for facilitating a fundraiser comedy night in support of this research, as well as the University of Guelph Hospitality Services for allowing a fundraiser bake sale to take place in the Summerlee Science Complex Atrium at the University of Guelph.
Author information
Authors and Affiliations
Contributions
KN, JM, HL, and BR proposed, secured funding for, conducted, and summarized this research in the present manuscript. KN completed the statistical analysis. TL-Q provided mentorship both in the field and in the lab, facilitated and completed qPCR of samples, significantly contributed to the methods section and provided invaluable feedback during manuscript production. DB assisted with field sampling, eDNA extraction, and creation of the qPCR standard fluorescent curve. RH provided lab and reagent access, organized partner meetings to help secure funding for this research, and acted as the principal investigator during the production of this manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All authors provide consent for transfer of non-exclusive publication rights and warrants that their contributions are original and that they have full power to make this grant.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nolan, K.P., Loeza-Quintana, T., Little, H.A. et al. Detection of brook trout in spatiotemporally separate locations using validated eDNA technology. J Environ Stud Sci 13, 66–82 (2023). https://doi.org/10.1007/s13412-022-00800-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13412-022-00800-x