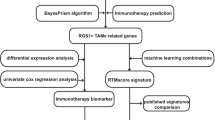
Abstract
Purpose
The tumor immune microenvironment (TME) plays a vital role in tumorigenesis, progression, and treatment. Macrophages, as an important component of the tumor microenvironment, play an essential role in antitumor immunity and TME remodeling. In this study, we aimed to explore the different functions of different origins macrophages in TME and their value as potential predictive markers of prognosis and treatment.
Methods
We performed single-cell analysis using 21 lung adenocarcinoma (LUAD), 12 normal, and four peripheral blood samples from our data and public databases. A prognostic prediction model was then constructed using 502 TCGA patients and explored the potential factors affecting prognosis. The model was validated using data from 4 different GEO datasets with 544 patients after integration.
Results
According to the source of macrophages, we classified macrophages into alveolar macrophages (AMs) and interstitial macrophages (IMs). AMs mainly infiltrated in normal lung tissue and expressed proliferative, antigen-presenting, scavenger receptors genes, while IMs occupied the majority in TME and expressed anti-inflammatory, lipid metabolism-related genes. Trajectory analysis revealed that AMs rely on self-renew, whereas IMs originated from monocytes in the blood. Cell-to-cell communication showed that AMs interacted mainly with T cells through the MHC I/II signaling pathway, while IMs mostly interacted with tumor-associated fibrocytes and tumor cells. We then constructed a risk model based on macrophage infiltration and showed an excellent predictive power. We further revealed the possible reasons for its potential prognosis prediction by differential genes, immune cell infiltration, and mutational differences.
Conclusion
In conclusion, we investigated the composition, expression differences, and phenotypic changes of macrophages from different origins in lung adenocarcinoma. In addition, we developed a prognostic prediction model based on different macrophage subtype infiltration, which can be used as a valid prognostic biomarker. New insights were provided into the role of macrophages in the prognosis and potential treatment of LUAD patients.
Similar content being viewed by others
1 Introduction
Lung cancer is the second most common cancer and the leading cause of cancer-related death worldwide [1], and the incidence of lung cancer has steadily increased over the past three decades [2]. Non-small cell lung cancer (NSCLC) accounts for nearly 85% of lung cancer, and lung adenocarcinoma (LUAD) is the most common histologic subtype of NSCLC, accounting for about 50%—70% of LUAD [3,4,5]. The tumor is a very heterogeneous tissue, and the tumor ecosystem consists of tumor cells, immune cells, stromal cells, and other cellular subtypes [6, 7]. The cell subtypes interact with each other through complex cellular communication and metabolites to promote tumor progression as well as response to or escape from therapy [8, 9]. With the application of new treatments such as targeted therapy and immunotherapy in lung cancer patients, the 5-year survival rate of advanced lung cancer has increased from less than 5% to about 30% [10, 11]. However, the biomarkers that can predict immunotherapy effects and the benefit population are still a barrier to the application of immunotherapy. Thus, further understanding of the tumor microenvironment (TME) can help us to improve personalized treatment strategies.
Tumor-associated macrophages (TAM) are usually one of the most infiltrated immune cell subtypes in solid tumors [12,13,14,15] and play an essential role in the regulation of tumor inflammation and angiogenesis [16, 17]. Usually, the activation status of TAM in vivo is divided into 'classically activated' M1 and 'alternatively activated' M2 [18]. However, TAM exhibits considerable heterogeneity in vivo, not only in different tumors but also in different patients or at different stages with the same tumor [19, 20]. Depending on the source, macrophages in the lung can be divided into alveolar macrophages (AMs), which are differentiated from embryonic erythromyeloid progenitor cells and fetal liver monocytes [21,22,23], and interstitial macrophages (IMs), which are derived from blood monocytes arising from hematopoietic stem cells in the bone marrow [24]. Mouse AMs develop from fetal monocytes after activation by the granulocyte–macrophage colony-stimulating factor (GM-CSF) and are highly expressed peroxisome proliferator-activated receptor gamma (PPARγ) [25, 26]. At the steady state, mouse AMs are maintained primarily by local self-renewal [27]. Interstitial macrophages are mainly derived from blood monocytes and are slowly renewed by circulating monocytes [28, 29].
Compared with traditional bulk RNA-sequencing, single-cell RNA-sequencing (scRNA-seq) can potentially probe tumor heterogeneity mechanisms and decode intercellular signaling networks, providing a new way for personalized therapy. In addition, it can identify the molecular characteristics of different infiltration immune cells in TME, providing new ideas for studying cancer immunity. Here, we compared the differences between AMs and IMs at the single-cell level in terms of gene expression, cellular communication, and differentiation. Subsequently, deconvolution to bulk RNA-seq data was performed for prognosis prediction of LUAD. This macrophage-based signature was further analyzed with mutation, metabolism, and immune cell infiltration to assess the association of macrophage signature with cancer immunity.
2 Materials and methods
2.1 Single-Cell RNA sequencing data and Bulk-RNA sequencing data collection
scRNA-seq was performed on 14 treatment-naive patients who underwent surgery at Zhongshan Hospital Fudan University with a postoperative pathological diagnosis of lung adenocarcinoma. In addition, seven normal and two LUAD samples and five normal samples were obtained from ArraryExpress (accession numbers EMTAB- 6149 and E-MTAB-6653) and Human Cell Atlas Data Coordination Platform (accession number PRJEB31843), respectively. And four tumors, normal lung tissue, and peripheral blood samples were from the GSE127465 dataset [30]. TCGA data and patient phenotypes were downloaded from the TCGA-LUAD dataset. The GEO data were obtained from four datasets, GSE50081, GSE37745, GSE31210, and GSE30219, respectively.
2.2 The scRNA dataset integration and cell annotation
Tissue dissected into single cells, suspension, and sequencing were described in our previous article [31]. After quality control, normalization, and PCA dimension reduction of scRNA-seq data using the Seurat package, data from different datasets were de-batched and normalized by the SCTransform package [32]. Cell clustering was based on PCA dimensionality reduction using the first 20 PCs and a resolution value of 0.2. FindAllMarkers was used to identify the marker genes for each subpopulation and subsequently annotate the cells.
2.3 Flow cytometry
Fresh lung cancer and normal tissues from 6 patients were collected, rinsed with ice-cold saline to remove extraneous blood. GentleMACS dissociator and gentleMACS C tubes (Miltenyi Biotec, Bergisch Gladbach, Germany) were used to dissociate the tissues into single cell suspensions by protocol. After cell counting, the cells were resuspended with PBS to a cell concentration of 1 × 107/mL. After blocking Fc receptors with Fc receptor blocker for 10 min, the supernatant was discarded and the cells were resuspended by 100μL PBS. Then cells and APC-Cy7-conjugated mouse anti-human LIVE/DEAD viability dye (5 μL/106 cells; BD Biosciences), FITC-conjugated mouse anti-human CD11b (5 μL/106 cells; Biolegend), PerCP-Cy5.5-conjugated mouse anti-human FABP4 (5 μL/106 cells; Abcam) and APC-conjugated rabbit anti-human SPP1 (5 μL/106 cells; Abcam) were incubated on ice for 1 h. Next, after centrifugation, the supernatant was discarded and resuspend the cell by PBS, FACSAria III (BD Biosciences) was applied to quantitate cells. Results were analyzed by FlowJo software (TreeStar, Woodburn, OR, USA).
2.4 Immunofluorescence assay
The steps for immunofluorescence are the same as shown in our previous article [33]. The paraffin-embedded slides were incubated with mouse anti-human CD11b (ab212505, Abcam), rabbit anti-human SPP1 (ab214050, Abcam), or rabbit anti-human FABP4 (ab92501, Abcam) and then horseradish peroxidase-conjugated secondary antibody. After that, the slides were stained with DAPI.
2.5 Pseudotime trajectory analysis for macrophages
To investigate the relationship between trajectories and macrophage subpopulations, we used the Monocle3 R package to construct macrophage differentiation trajectories after specifying the corresponding cells as root nodes. Subsequently, graph_test was used to find the pseudotime trajectory difference genes, and the obtained genes were used to plot the heat map.
2.6 Cell-to-cell communication analysis
First, we constructed Cell Chat objects from the Seurat data and used the CellChatDB.human database to infer the ligand-receptor cellular communication network. netVisual_circle function showed the strength of the communication network from different macrophages to other different cell types in the TME. netVisual_bubble function showed a bubble map of significant ligand-receptor interactions between macrophages and other cell types.
2.7 The CIBERSORTx analysis
First, we used our single-cell data and CIBERSORTx (https://cibersortx.stanford.edu) to construct signature genes for different cell types and subsequently calculated TCGA cell type abundance based on the matrix of signature genes [34].
2.8 Construction and validation of prognostic model
We used the glmnet package to perform LASSO Cox regression analysis to investigate the infiltration of macrophage subpopulations in relation to overall survival (OS) in TCGA-LUAD patients and to construct a risk model. Multifactorial Cox regression analysis was then used to determine the significance of the risk model versus clinical phenotype on prognosis. pROC package was used to predict the model's receiver operating characteristic (ROC) curve.
2.9 Statistical analysis
All statistical analyses were performed by R version 4.0.2. Continuous variables were assessed by an independent Student's t-test. Non-normally distributed data were analyzed using the Wilcoxon rank sum test. Categorical variables were analyzed by chi-square test or Fisher's exact test. The relationship between gene expression levels was assessed based on Spearman's correlation coefficient. Two-sided p < 0.05 was the threshold of significance. The threshold for differential genes was defined as | logFC |> 1, adj. p < 0.05. *P < 0.05, **P < 0.01, ***P < 0.001.
2.10 Recognition of different TTK patterns by unsupervised clustering
NMF decomposition was used to identify potential mutation patterns in risk group patients using the method of Hong et al. using the TCGA-LUAD cohort somatic cell mutation data for analysis [35]. To further explore the etiology of patients in different risk subgroups, mutation patterns were compared with mutation features from the COSMIC database, and each mutation pattern was annotated.
3 Results
3.1 Single-cell expression profiling of myeloid cells and macrophages in LUAD
In order to clarify the role of myeloid cells and macrophages in the development of LUAD, we performed single-cell RNA sequencing using tumor tissues from 13 treatment-naive early and 8 advanced LUAD patients, 12 normal tissues, and four peripheral blood data. The baseline information of these patients is available in Supplementary Table 1. After dimension and clustering, we extracted myeloid cells from different datasets according to their common markers, such as LYZ, CD68, and CD1C, for subsequent studies (Supplementary Fig. 1a & b). A total of 24062 myeloid cells were obtained, including 13,783 cells from tumor tissues, 10,065 cells from normal tissues, and 217 from peripheral blood respectively (Fig. 1a). After removing batch effects using the SCTransform package to combine data from different datasets, we obtained 10 clusters for further study (Supplementary Fig. 1c & 2a). In these 10 clusters, differentiation of monocytes from macrophages is based on the presence of monocyte markers (FCN1, LYZ, VCAN) and the absence of macrophage markers (CD68, GPNMB, CTSB). Therefore, we defined clusters 4 and 9 as monocytes and clusters 0, 1, 5, 6, 7, and 8 as macrophages. Besides the characteristic macrophage markers MARCO and MRC1, cluster 0 and 6 expressed alveolar resident cell-associated markers (FABP4, PPARG) and cell cycle genes (MKI67, STMN1), which are consistent with the clearance of surface-active proteins and self-renewal potential of alveolar-resident macrophages, and thus were defined as alveolar macrophages (AMs) and clusters 1, 5, 7, and 8 were defined as interstitial macrophages (IMs). In addition, we also found that cluster 2 and 3 expressed monocyte makers as well as macrophage makers, so we defined them as intermediate stages in the evolution of monocytes to macrophages, mo-Mac (Fig. 1b, c & Supplementary Fig. 2b).
Annotation of myeloid cells and macrophages by scRNA-seq in tumor, normal tissue, and peripheral blood a) UMAP plot colored by cells of different origins b) UMAP plot of marker gene for four cell types (from left to right: monocyte, AM, IM) c) UMAP plot colored by different clusters d) Relative proportion of cell subsets for different tissue origins e) Relative contribution of each cell type in normal vs. tumor tissue and early vs. advanced LUAD f) Difference genes between AMs and IMs, with a threshold of |log2FC|> 2, |Difference|> 0.2, up represents genes highly expressed in IMs, and down indicates genes highly expressed in AMs g) GSVA analysis of differential genes between AMs and IMs, up represents enriched pathways in AMs, and down is enriched in IMs
Quantitatively, peripheral blood was mainly composed of monocytes, whereas IMs and AMs infiltration in normal and tumor tissues were mainly present (Fig. 1d). Compared to normal tissue, IMs and mo-Mac infiltration were increased in tumors, while the number of AMs and monocytes was decreased. AMs were similarly reduced in advanced LUAD patients, whereas the other three subtypes of myeloid cells were altered, but not significantly (Fig. 1e).
We next compared the differences in gene expression between AMs and IMs (Fig. 1f), and GSVA analysis showed that AMs were mainly associated with neurological processes, whereas IMs were related to metabolic (Fig. 1g).
3.2 Phenotypic heterogeneity of myeloid cells and macrophages in LUAD
Based on the markers expressed in each cluster, we continued to classify monocytes into CD14 + monocytes (those expressing classical monocyte markers, like IL1RN and S100A8/9), and non-classical CD16 + monocytes (highly expressing Leukocyte Immunoglobulin Like Receptor (LILR) family). The mo-Mac cells can also be divided into IL1B + mo-Mac and HSPA6 + mo-Mac, characterized by the expression of IL1B, CXCL8, CXCL3, or heat-shock proteins (HSPA6 and HSPA1A/B), respectively. IMs could be further delineated into SPP1 + , CCL18 + , IFITM3 + , and MT1G + macrophages based on the expression of different characteristic genes. SPP1 + macrophages mainly highly express SPP1, GPNMB, CSTB, and other oncogenes, and several studies have shown their relationship with tumor progression. CCL2, CCL18, and other chemokines characterized CCL18 + macrophages. In contrast, IFITM3 + macrophages mainly express IFN-induced antiviral genes, which may have an important role in anti-infection. In addition, MT1G + macrophages were identified by high amounts of metallothionein, suggesting an association with oxidative stress and apoptosis. Finally, we defined cluster 0 as FABP4 + Alveolar-mac and cluster 6 as STMN1 + Alveolar-mac (Fig. 2a & Supplementary Fig. 2c).
Diversity within the myeloid cells and macrophages lineage and functionality according to tissue origins a) Complex heatmap of selected marker genes in each cell cluster. Up: Tissue preference of each cluster. Down: Relative expression of marker genes associated with each cell subset b) Heatmap showing myeloid cell and macrophage phenotypes with corresponding functional genes c) Heatmap showing myeloid cell and macrophage phenotypes with corresponding functional gene sets and pathways
After that, we divided the common cytokines of myeloid cells and macrophages into pro-inflammatory, anti-inflammatory gene sets, and antigen-presentation-related MHC-I/II genes and investigated their expression in different clusters. We found that monocytes and IL1B + mo-Mac mainly expressed pro-inflammatory genes. HSPA6 + mo-Mac, in addition to expressing pro-inflammatory genes, also expressed anti-inflammatory factors such as CCL13, FLOR2, and receptor-mediated phagocytosis genes such as MERTK, suggesting an intermediate state in the evolution of monocytes to macrophages. Among IMs, SPP1 + and CCL18 + macrophages mainly expressed anti-inflammatory factors, while IFITM3 + macrophages highly expressed anti-inflammatory and pro-inflammatory factors such as CCL2 and CCL3. MT1G + macrophages expressed low in both pro- and anti-inflammatory factors and only expressed a small number of receptor-mediated phagocytosis genes. On the other hand, Alveolar-resident macrophages predominantly express major histocompatibility complex (MHC) related genes, suggesting a key role in antigen presentation and intrinsic immunity (Fig. 2b). In terms of pathways, IMs were associated with anti-inflammatory and IL2-STAT5, IL6-JAK2-STAT3 pathways. AMs were related to inflammation and interferon genes (Fig. 2c).
We observed a significant increase in SPP1 + , CCL18 + , IFITM3 + macrophages, and STMN1 + alveolar macrophages in tumor tissues verse normal tissues. Vice versa, FABP4 + alveolar macrophages, and IL1B + mo-Mac were reduced in the tumor microenvironment. FABP4 + alveolar macrophages infiltration reduced both in the early and advanced tumor comparison, albeit non-significantly. IFITM3 + macrophages and HSPA6 + mo-MAc infiltration were increased in advanced LUAD, suggesting a corresponding relationship in tumor progression, while IL1B + mo-Mac infiltration was reduced (Fig. 3a, b & Supplementary Fig. 2d). Subsequently, we selected SPP1 + and FABP4 + macrophage, two of the more predominant macrophages, for validation in our patient samples. The results of flow cytometry revealed a significantly higher macrophage infiltration in the tumor compared to normal tissue. In addition, SPP1 + macrophage infiltration was increased in tumor samples, whereas in normal tissues it was predominantly FABP4 + macrophages (Fig. 3c & Supplementary Fig. 2e). Similarly, the results of our immunofluorescence assay also confirmed this conclusion (Fig. 3d).
Infiltration of different macrophage subtypes a) Relative contribution of each cell type in normal vs. tumor b) Relative contribution of each cell type in early vs. advanced LUAD c) Proportion of SPP1 + macrophage and FABP4 + macrophage in tumor and normal tissue macrophages d) Immunofluorescence shows the infiltration of SPP1 + macrophage and FABP4 + macrophage in tumor and normal tissues
3.3 The trajectory of macrophages differentiation in LUAD
Subsequently, we reconstructed the cellular lineages of differentiation trajectories of macrophages using pseudotime analysis. After using blood monocytes as the starting point, we can see that monocytes in the blood were first transformed into monocytes in the lung and confluence branch into mo-Mac, the latter continuing to transit into monocyte-derived macrophages (IMs). We can also see no obvious relationship between AMs and monocyte populations, suggesting their self-renewal function (Supplementary Fig. 3a). Therefore, we divided them into two categories: AMs and non-AMs (including monocytes, mo-Mac, and IMs). We performed re-clustered and trajectory analysis in the above two categories. Similarly, we found that monocytes in the blood first differentiate into mo-Macs, since they send branches to individual macrophages (Fig. 4a Supplementary Fig. 3b). Alveolar macrophages, on the other hand, rely mainly on self-renewal (Fig. 4b). Moreover, the density plot also revealed that macrophages from tumor tissues were mainly enriched in the latter half of the linear, which confirmed our above finding of IMs enrichment in the tumor tissues (Supplementary Fig. 3c).
The trajectory of macrophage differentiation in LUAD a) Pseudotime trajectories for IMs based on monocle3 b) Pseudotime trajectories for AMs based on monocle3 c) Gene expression dynamics along the IMs lineage d) Profiling of M1-like or M2-like signaling along the IMs lineage e) Gene expression dynamics along the AMs lineage
Modeling gene expression along the IMs lineage revealed five gene sets (Fig. 4c). Sets 1 mainly expresses monocyte-associated genes, and inflammatory markers, suggesting the differentiation started from monocytes and the inflammatory state of monocytes. Set 2 and 3 were characterized by transcription factors, inflammatory markers, heat-shock proteins, and metallothionein in the lineage. It indicated that monocytes and mo-Mac were in a pro-inflammatory and oxidative stress state, which may suggest their potential anti-cancer effects in TME. Notably, in set 3, we can see a high expression of SPP1, which induces differentiation of macrophages to tumor-associated macrophages, promotes angiogenesis and tumor metastasis, and is associated with a worse prognosis. Furthermore, we also observed set 5 was expressed at the end of the trajectory and comprised genes involved in anti-inflammatory, lipid degradation or metabolism genes, and genes associated with macrophage pro-oncology (LGMN, CSTB, GPNMB), suggesting that they may play a key role in promoting tumor progression. In addition, we compared the traditional classical or alternative activation pathway (M1/M2) of macrophages and pro-inflammatory or anti-inflammatory functions with pseudotime. We found that classical pathway (M1-like) was highly expressed at the beginning and then decreased, while alternative activation pathway (M2-like) was gradually increased. Similar results were observed for pro-inflammatory and anti-inflammatory functions (Fig. 4d & Supplementary Fig. 3d).
Modeling gene expression along the alveolar macrophages’ lineage revealed three gene sets (Fig. 4e). Set1 mainly expressed cell proliferation-related genes (MKI67, TOP2A) and transcription factors (CDK1, PLK1), suggesting its high proliferative activity. Sets 2 and 3 comprised genes involved in receptor-mediated phagocytosis, such as scavenger receptors MARCO and MSR1, complement activation (C1QA, C1QB, C1QC), antigen presentation (HLA-DRA, HLA-DRB1, HLA-DPA1, HLA-DPB1), as well as anti-inflammatory markers (PPARG, FABP4). This cluster cells were similar to the main expression phase of FABP4 + alveolar macrophages, indicating that this cluster has the function of antigen presentation, activation of innate immunity, and thus possible tumor killing.
3.4 Cell-to-cell communication to unravel the macrophage context in LUAD
Subsequently, we explored and predicted the interactions between IMs and AMs with other cells in the TME. First, we calculated the interactions between different types of macrophages and other cells, and assessed the interaction strength (Supplementary Fig. 4a). In addition to their self-interaction, the interactions between all kinds of macrophages with T cells were the most obvious and frequent. Meanwhile, the interaction between IMs with fibroblasts and tumor cells was also more significant than in other macrophage types. For alveolar macrophages, their interactions with other cells are almost always involved in major histocompatibility complexes and antigen presentation (MHC-I and MHC-II signaling pathways), phagocytosis (MARCO, MRC1), and FN1 or CD44-rich extracellular matrix, such as FN1-CD44, COL-CD44 (Fig. 5a). IMs interactions were mainly about migratory and proliferative effects, especially with endothelial, epithelial, and cancer cells (CXCL8-ACKR1). Interstitial macrophages also presented antigen to T cells but were not as strong as alveolar macrophages. Notably, the interaction of the SPP1 pathway between monocyte-derived macrophages with the immune cell was also significantly enhanced (SPP1-CD44) (Fig. 5a). The interactions between MRC1, secreted by AMs and PTPRC, especially expressed on the surface of T cells were enhanced. MRC1 plays a key role in myeloid plasticity and can improve adaptive immune responses. IMs, on the other hand, interact more closely with stromal cells, especially based on the SDC4-COL family (Fig. 5b). Finally, we selected four pathways we were interested (SPP1, FN1, MHC-I, and MHC-II) to identify the role of each type of cell in them (Supplementary Fig. 4b). It can be seen that macrophages mainly acted as senders in the MHC pathway, and alveolar macrophages were of higher importance. In contrast, in the FN1 pathway, alveolar macrophages act primarily as regulators, regulating signals sent from fibroblasts to other cells. On the other hand, interstitial macrophages affected other cells mainly through the secretion of SPP1. Notably, the downstream influencers of the SPP1 pathway were mostly immune cells, with almost no effect on tumor cells and fibroblasts.
3.5 Macrophage infiltration contributed the LUAD prognosis and immunotherapy
Then, we mapped our resulting single-cell data to TCGA transcriptome data using the deconvolution approach to calculate the percentage of different subtypes of macrophages. Similarly, the infiltration of alveolar macrophages and mo-Mac was significantly higher in normal tissues, while monocyte-derived macrophages and monocytes were less frequent (Supplementary Fig. 5a). As for subgroups, FABP4 + Alveolar Mac and IL1B + mo-Mac were more infiltrated in normal tissues, while SPP1 + Macrophage, STMN1 + Alveolar Mac, and IFITM3 + Macrophage were increased in tumor tissues. In terms of macrophage infiltration between early stage and advanced LUAD, monocyte was slightly increased, and no significant difference between the rest (Supplementary Fig. 5b).
To identify prognostic features based on macrophage infiltration, we used the LASSO Cox regression model to calculate the relationship between the different subpopulation macrophage infiltration and prognosis. Subsequently, a risk score was established: risk score = (3.02 × Monocyte expression) + (-2.06 × mo-Mac expression) + (1.73 × Macrophage expression) + (-1.20 × Alveolar mac expression). The risk score was calculated for each patient using this formula. Patients were divided into a high-risk group (n = 251) and a low-risk group (n = 251) based on the median cutoff (cutoff value = 1.957), and the distribution of risk scores and survival status for each patient were shown in Fig. 6a. Baseline information of overall patients and high and low-risk subgroups was presented in supplementary Table 2. The heat map shows the specific level of infiltration of the four types of myeloid cells or macrophages (Supplementary Fig. 5c). Kaplan–Meier survival analysis showed that overall survival was significantly higher in low-risk patients than in high-risk patients (P = 0.0041) (Fig. 6b). We assessed the predictive accuracy of the prognostic features using time-dependent ROC curves for OS at 2, 3, and 5 years (Fig. 6c). The AUC values at these times were 0.719 (95% CI: 0.667—0.771), 0.717 (95% CI: 0.668—0.767), and 0.675 (95% CI: 0.626—0.724), respectively. Univariate and multivariate Cox regression analyses were performed to further explore whether the risk score was an independent factor in LUAD patients. The results of multivariate cox regression models confirmed that risk score was a significant factor independent of age, sex, and clinical symptoms (HR: 6.764, 95% CI: 2.312 – 19.789, p < 0.001) (Supplementary Table 3).
Construction of prognostic signature based on macrophage infiltration in TCGA and GEO database a) The distribution of risk scores and survival status of the TCGA patients b) Kaplan–Meier curves of survival analysis in TCGA LUAD patients based on the risk score c) ROC analysis of the predictive model for predicting the risk of death at 2, 3, and 5 years in the TCGA database d) The distribution of risk scores and survival status of the GEO patients e) Kaplan–Meier curves of survival analysis in GEO LUAD patients based on the risk score f) ROC analysis of the predictive model for predicting the risk of death at 2, 3, and 5 years in the GEO database
To validate the accuracy of our risk score in predicting prognosis, we used four different GEO datasets (GSE50081, GSE37745, GSE31210, GSE30219) and selected LUAD patients after a de-batch merge. The percentage of different macrophages was calculated by deconvolution, and then the risk score was calculated for each patient using the above formula. Similarly, patients were divided into low-risk (n = 271) and high-risk groups (n = 273) based on median cutoff values (cutoff value = -0.968) (Fig. 6d). Baseline information was demonstrated in supplementary Table 4. Kaplan–Meier survival analysis showed that patients in the low-risk group had significantly better OS than those in the high-risk group (p = 0.0014) (Fig. 6e). In addition, the accuracy of the predictive assessment was confirmed by ROC curves using 2-, 3-, and 5-year OS. The AUC values for these times were 0.689 (95% CI: 0.634—0.743), 0.647 (95% CI: 0.594—0.700) and 0.626 (95% CI: 0.577–0.675), respectively (Fig. 6f). Multivariate cox regression models also showed that risk score was a significant factor independent of age, gender, and tumor stage (HR: 1.816, 95% CI: 1.324 – 2.492, p < 0.001) (Supplementary Table 5).
In addition, we observed the same significance of macrophage infiltration in predicting immune response in eight public cancer cohorts receiving immunotherapy. These included clear cell renal cell carcinoma (ccRCC), non-small cell lung cancer (NSCLC), melanoma, met-melanoma, and urothelial cancer (Fig. 7a).
Genomic features and signaling pathways associated with the high and low-risk groups a) Macrophage infiltration contributed on immunotherapy b) Differential genes between the high-risk and low-risk groups with a threshold of |log2FC|> 1, adj. p.value < 0.05, up represents genes highly expressed in high-risk group, and down indicates genes highly expressed in low-risk group c) GSVA analysis of differential genes between high and low-risk groups d) The 22 immune cells' infiltration between high-risk and low-risk groups
3.6 Differences in gene expression and immune cell infiltration between high and low-risk groups
Next, we performed differential gene analysis for the high- and low-risk groups and obtained a total of 138 up-regulated and 219 down-regulated genes in the high-risk group (Fig. 7b). By enrichment analysis, we found those differential genes were mainly enriched in antigen presentation, immunomodulatory, and extracellular matrix-related pathways related to the biological function of macrophages (Supplementary Fig. 6). GSVA analysis showed that the high-risk group was mainly enriched in metabolism-related pathways such as alditol metabolic process and disulfide oxidoreductase activity, while the low-risk group was mostly associated with antigen presentation, immune activation pathways, such as 'antigen processing and presentation of endogenous lipid antigen via MHC class IB', and 'activated T cell proliferation' (Fig. 7c). Interestingly, some of the pathways enriched in the low-risk group were associated with microglia. Microglia are a specialized type of macrophage located in the brain and spinal cord and, similar to alveolar macrophages, are embryonic tissue-resident macrophages, responsible for the immune defense of the central nervous system [36].
Since macrophages play the phagocytic, antigen presentation role in antitumor immunity, we explored the relationship between high and low-risk groups and immune cell infiltration in patients with LUAD. CIBERSORT analysis showed a high proportion of macrophage M0 and macrophage M2 in the high-risk group, while a high infiltration of B cells memory, dendritic cells resting, mast cells resting, T cells CD4 memory activated, T cells CD4 memory resting, T cells CD8 in the low-risk group (Fig. 7d).
3.7 Genomic features and signaling pathways associated with the high and low-risk groups
We analyzed the distribution of somatic mutations among the two groups using mutation data from the TCGA dataset. The frequency of mutations was significantly higher in the high-risk group compared to the low-risk group (Fig. 8a). Common mutations in lung cancer, such as TP53, TTN, and CSMD3 mutations, were predominant in the high-risk group. Since somatic mutations result from a combination of factors, different mutational processes lead to different mutation types or characteristics. Therefore, we identified three mutational features for both groups of patients, which were used to represent the mutational characteristics comprehensively (Fig. 8b). Mutations in the high-risk group were mainly associated with signatures 4, 6, and 13, and in the low-risk group with signatures 1, 4, and 13. Signature 1 is the result of an endogenous mutational process initiated by spontaneous deamination of 5-methylcytosine. Signature 4 is associated with smoking and is likely due to tobacco mutagens. Signature 6 is associated with defective of DNA mismatch repair and is found in unstable microsatellite tumors. And signature 13 has been attributed to the activity of the AID/APOBEC family of cytidine deaminases converting cytosine to uracil. Smoking-induced mutations dominated the high-risk group, while the AID/APOBEC family was the predominant pattern in the low-risk group (Fig. 8c). The above study suggests that although the somatic mutation genes were similar in the high and low-risk groups, there were still differences in mutation rates and factors, which was one of the potential factors that may contribute to survival differences.
Somatic mutations and mutational signatures in the high and low-risk groups a) Waterfall plots revealing the somatic mutation distributions with the highest mutation frequencies in the high (left) and low-risk (right) groups b) Bayesian non-negative matrix factorization was used to determine the optimal number of mutational signatures, and based on 30 mutation features summarized in the COSMIC database, three signatures for the two clusters were annotated c) Upper panel, Bayesian NMF was used to identify three mutational signatures in the high (left) and low-risk (right) groups
4 Discussion
Using sc-RNA data from tumor tissue, normal tissue, and peripheral blood samples from LUAD patients, we performed an in-depth analysis of macrophages in the LUAD TME depending on their origin. Notably, our study incorporated monocyte cells from peripheral blood of tumor patients and used it as a starting point for the pseudotime analysis of macrophage differentiation, which can more accurately determine the origin and differentiation trajectory of different types of macrophages. In contrast to the relatively stable phenotype of lymphocytes and stromal cells in different tissues and cancer types, the different characteristics of macrophages seem to depend on their tissue origin [30, 37, 38]. Although the previous study showed that TAM in lung cancer exhibits a continuous spectrum phenotype [30], we found that alveolar and interstitial macrophages express different phenotypic and functional characteristics. In this study, we performed differential gene analysis, cell-to-cell communication, and pseudotime analysis between alveolar macrophages and interstitial macrophages. Markers of different subtypes of macrophages were then projected onto the bulk RNA-seq data of LUAD to construct a new prognostic predictive signature, which was identified as an independent risk factor for LUAD patients. Transcriptomic, metabolic, immune infiltration and mutational analyses were also performed on the high and low-risk groups to assess potential reasons for survival differences between the two groups.
Firstly, we found that in normal tissues, alveolar macrophages, especially FABP4 + AMs, were more predominant, whereas IMs were more infiltrated in tumors, such as SPP1 + , CCL18 + , IFITM3 + macrophages. AMs are mainly characterized by their anti-inflammatory, pro-phagocytic, and antigen presentation functions, suggesting their role in clearing dead alveolar epithelial cells and foreign pathogenic bacteria in normal lung tissue, and clearing tumor cells in tumors, which is consistent with the previously reported function of alveolar macrophages [39]. In inflammatory diseases, AMs' function is thought to contribute to disease amelioration and inflammation regression [40]. FABP4 + macrophages play an important role in infection. Studies have shown that their infiltration increases in mild COVID-19 patients [41]. In addition, it can recruit neutrophils and clear pathogenic bacteria. After knocking down FABP4, P. aeruginosa infection mice exhibited reduced bacterial clearance and increased mortality [42]. In tumors, Zhou Y et al. found increased infiltration of FABP4 + macrophages in metastatic lung osteosarcoma [43]; however, its role in lung cancer needs further study. Interstitial macrophages are predominantly characterized by the expression of anti-inflammatory-related genes, and previous studies have shown that they are essential for maintaining lung homeostasis [28, 44]. SPP1 + macrophages are considered tumor-specific macrophages, which are highly expressed in a variety of tumor tissues and hardly expressed in normal tissues, exerting pro-angiogenic and pro-tumor metastatic functions, and high infiltration levels are associated with worse prognosis [45,46,47]. In lung cancer, SPP1 + macrophages were found to mediate macrophage polarization, lung cancer immune evasion, and angiogenesis [48, 49], and may be a novel biomarker for immunotherapy [50]. Similarly, CCL18 + macrophages have been found to exhibit extremely high metabolic activity in the TME and are associated with a poorer prognosis [51, 52]. In contrast, effective neoadjuvant chemotherapy can downregulate this metabolic activation and offer the possibility of targeting metabolic pathways for treatment [53].
Secondly, the results of the pseudotime analysis showed that interstitial macrophages mainly originated from monocytes in the blood, especially CD14 + monocytes, whereas alveolar macrophages were independent of monocytes and mainly self-renewed. During the evolution from monocytes to interstitial macrophages, we found a shift in gene expression from monocyte-associated genes, pro-inflammatory genes to heat shock proteins, metallothionein, and finally to anti-inflammatory genes and genes associated with pro-carcinogenesis [54,55,56]. This trajectory reflects the continuous process of macrophage transition from anti-cancer to pro-cancer, suggesting the heterogeneity of macrophages in TME. Macrophages as a double-edged sword. How to utilize their anti-cancer role more efficiently and inhibit their pro-cancer is the key to recent research. As for alveolar macrophages, there is a shift from proliferation genes to phagocytosis, MHC genes, and complement components activating functional genes. This suggests their role in antigen presentation and activation of adaptive immunity [57,58,59]. Maximal heterozygosity at HLA-I loci can improve the overall survival of immunotherapy compared with patients who were homozygous for at least one HLA locus [60]. This may be related to its strong antigen-presenting ability, which makes it more likely to benefit from immunotherapy. In cell-to-cell communication, alveolar macrophages mainly interact with CD4 + T cells and CD8 + T cells through the MHC pathway, whereas interstitial macrophages mainly interact with fibroblasts and tumor cells by SPP1. Cancer-associated fibroblasts (CAFs) and TAMs usually interact through the CSF1-CSF1R axis [61]. Zhang et al. showed that SPP1 + TAM is more likely to interact with CAFs through syndecan-2 expression on TAM, which binds to matrix metallopeptidase 2 (MMP2) on CAFs [47]. In subcutaneous or in situ mouse models of hepatocellular carcinoma (HCC), SPP1 + tumors responded better to combined treatment with PD-L1 blockade and CSF1R inhibition [62]. This may be related to the fact that a relatively high proportion of immunosuppressive TAM (expressing SPP1) was depleted by CSF1R inhibitors, leading to a higher degree of TME remodulation, favoring tumor suppression. In addition to fibroblasts, CSF1 can also be secreted by tumor cells, suggesting a potential pro-tumor effect [63]. Apart from CSF1-CSF1R signaling, CAFs can also recruit monocytes and macrophages into TME via CCL2-CCR2, the CXC chemokine family [64,65,66]. Knocking down the CCL2 gene in CAFs, thus blocking the CCL2-CCR2 axis, can inhibit tumor growth [67].
The predictive power of the prediction models based on the level of infiltration of different subtypes of monocytes/macrophages was well validated in both TCGA and GEO databases. Its powerful predictive power prompted us to explore the underlying mechanisms. Differential genetic analysis, GO analysis, and GSVA analysis of the high and low groups showed that the low-risk group was mainly associated with immune and antigen presentation, while the high-risk group was associated with metabolism. Tumor progression is often accompanied by metabolic reprogramming in order to meet the increased bioenergetic and biosynthetic demands. And in TME, the metabolism of other cell types besides tumor cells can also regulate tumor progression [68]. Tumor cells produce large amounts of lactate through the Warburg effect and release it into the extracellular microenvironment. Lactate can act as a messenger between tumor cells and TAM, inducing vascular endothelial growth factor (VEGF) expression and M2-like polarization [69]. In addition, other tumor metabolites, such as glutamine, succinate, and adenosine, can also induce TAM differentiation to the M2 phenotype and promote tumor progression [70,71,72]. Gene mutations play a key role in causing abnormal and uncontrolled growth of tumor cells. These mutations are known as "drivers" in driving tumorigenesis, conferring certain selective advantages to mutant cells over neighboring cells [73]. Different factors often cause gene mutations, and it is important to identify the various factors for us to take different interventions. Mutational signatures in the high-risk group were mainly identified to be associated with smoking. This may be related to the ability of smoking to recruit blood monocytes to differentiate into macrophages, producing a higher number of immature macrophages, promoting pro-inflammatory effects, and reducing phagocytosis and efferocytosis ability [74, 75]. The mutational signature of the low-risk group is associated with the AID/APOBEC family, a group of cytidine deaminases that deaminate cytidine to uridine and can therefore insert mutations in DNA and RNA. In the immune system, AIDs and APOBEC3 play a role in antigen-driven antibody diversification and innate defense systems against retroviruses, respectively. Also, given their ability to mutate DNA, they may have a potential role in cancer development [76].
The present study has several limitations. First, our study is only based on the single cell and public database and does not include any in vitro or in vivo experiments, which are needed to further explore the potential molecular mechanisms of macrophages in predicting prognosis and tumor progression. In addition, the predictive power of the model needs to be further validated in large-scale prospective clinical studies.
5 Conclusion
In conclusion, we investigated the composition, expression differences, and phenotypic changes of macrophages from different origins in lung adenocarcinoma. The functional transformation was also explored through pseudotime trajectories and cell-to-cell interactions. In addition, we developed a prognostic prediction model based on different macrophage subtype infiltration, which can be used as a valid prognostic biomarker. New insights were provided into the role of macrophages in the prognosis and potential treatment of LUAD patients.
Data availability
Single-cell data after integration are available upon request to corresponding authors. The TCGA database data is downloaded from http://portal.gdc.cancer.gov/. The GEO database data were integrated from four datasets, GSE50081, GSE37745, GSE31210, and GSE30219.
References
H. Sung, J. Ferlay, R.L. Siegel et al., Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries[J]. CA Cancer J Clin 71(3), 209–249 (2021). https://doi.org/10.3322/caac.21660
T. Lu, X. Yang, Y. Huang et al., Trends in the incidence, treatment, and survival of patients with lung cancer in the last four decades[J]. Cancer Manag. Res. 11, 943–953 (2019). https://doi.org/10.2147/CMAR.S187317
H. Asamura, T. Goya, Y. Koshiishi et al., A Japanese lung cancer registry study: prognosis of 13,010 resected lung cancers[J]. J. Thorac. Oncol. 3(1), 46–52 (2008). https://doi.org/10.1097/JTO.0b013e31815e8577
H. Nakamura, H. Saji, Worldwide trend of increasing primary adenocarcinoma of the lung[J]. Surg. Today 44(6), 1004–1012 (2014). https://doi.org/10.1007/s00595-013-0636-z
U. Testa, G. Castelli, E. Pelosi, Lung cancers: molecular characterization, clonal heterogeneity and evolution, and cancer stem cells[J]. Cancers (Basel) 10(8) (2018). https://doi.org/10.3390/cancers10080248
M. Binnewies, E.W. Roberts, K. Kersten et al., Understanding the tumor immune microenvironment (TIME) for effective therapy[J]. Nat. Med. 24(5), 541–550 (2018). https://doi.org/10.1038/s41591-018-0014-x
F. Wu, J. Fan, Y. He et al., Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer[J]. Nat. Commun. 12(1), 2540 (2021). https://doi.org/10.1038/s41467-021-22801-0
D. Chen, X. Zhang, Z. Li et al., Metabolic regulatory crosstalk between tumor microenvironment and tumor-associated macrophages[J]. Theranostics 11(3), 1016–1030 (2021). https://doi.org/10.7150/thno.51777
A. Ostman, The tumor microenvironment controls drug sensitivity[J]. Nat. Med. 18(9), 1332–1334 (2012). https://doi.org/10.1038/nm.2938
E. Nadal, B. Massuti, M. Domine et al., Immunotherapy with checkpoint inhibitors in non-small cell lung cancer: insights from long-term survivors[J]. Cancer Immunol. Immunother. 68(3), 341–352 (2019). https://doi.org/10.1007/s00262-019-02310-2
Z. Hu, M. Li, Z. Chen et al., Advances in clinical trials of targeted therapy and immunotherapy of lung cancer in 2018[J]. Transl. Lung Cancer Res. 8(6), 1091–1106 (2019). https://doi.org/10.21037/tlcr.2019.10.17
L. Cassetta, S. Fragkogianni, A.H. Sims et al., Human tumor-associated macrophage and monocyte transcriptional landscapes reveal cancer-specific reprogramming, biomarkers, and therapeutic targets[J]. Cancer Cell 35(4), 588–602 (2019). https://doi.org/10.1016/j.ccell.2019.02.009
J. Wagner, M.A. Rapsomaniki, S. Chevrier et al., A single-cell atlas of the tumor and immune ecosystem of human breast cancer[J]. Cell 177(5), 1330–1345 (2019). https://doi.org/10.1016/j.cell.2019.03.005
S. Chevrier, J.H. Levine, V. Zanotelli et al., An immune atlas of clear cell renal cell carcinoma[J]. Cell 169(4), 736–749 (2017). https://doi.org/10.1016/j.cell.2017.04.016
Z. Chen, M. Zhao, J. Liang et al., Dissecting the single-cell transcriptome network underlying esophagus non-malignant tissues and esophageal squamous cell carcinoma[J]. EBioMedicine 69, 103459 (2021). https://doi.org/10.1016/j.ebiom.2021.103459
M.C. Schmid, J.A. Varner, Myeloid cells in the tumor microenvironment: modulation of tumor angiogenesis and tumor inflammation[J]. J. Oncol. 2010, 201026 (2010). https://doi.org/10.1155/2010/201026
C. Engblom, C. Pfirschke, M.J. Pittet, The role of myeloid cells in cancer therapies[J]. Nat. Rev. Cancer 16(7), 447–462 (2016). https://doi.org/10.1038/nrc.2016.54
L. Cassetta, J.W. Pollard, Targeting macrophages: therapeutic approaches in cancer[J]. Nat. Rev. Drug Discov. 17(12), 887–904 (2018). https://doi.org/10.1038/nrd.2018.169
F. Ginhoux, J.L. Schultze, P.J. Murray et al., New insights into the multidimensional concept of macrophage ontogeny, activation and function[J]. Nat. Immunol. 17(1), 34–40 (2016). https://doi.org/10.1038/ni.3324
S. Cheng, Z. Li, R. Gao et al., A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells[J]. Cell 184(3), 792–809 (2021). https://doi.org/10.1016/j.cell.2021.01.010
P.E. Gomez, K. Klapproth, C. Schulz et al., Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors[J]. Nature 518(7540), 547–551 (2015). https://doi.org/10.1038/nature13989
G. Hoeffel, J. Chen, Y. Lavin et al., C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages[J]. Immunity 42(4), 665–678 (2015). https://doi.org/10.1016/j.immuni.2015.03.011
M. Guilliams, I. De Kleer, S. Henri et al., Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF[J]. J. Exp. Med. 210(10), 1977–1992 (2013). https://doi.org/10.1084/jem.20131199
E.G. Perdiguero, F. Geissmann, The development and maintenance of resident macrophages[J]. Nat. Immunol. 17(1), 2–8 (2016). https://doi.org/10.1038/ni.3341
C. Schneider, S.P. Nobs, M. Kurrer et al., Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages[J]. Nat. Immunol. 15(11), 1026–1037 (2014). https://doi.org/10.1038/ni.3005
F. Li, K.M. Okreglicka, L.M. Pohlmeier et al., Fetal monocytes possess increased metabolic capacity and replace primitive macrophages in tissue macrophage development[J]. EMBO J. 39(3), e103205 (2020). https://doi.org/10.15252/embj.2019103205
R. Gentek, K. Molawi, M.H. Sieweke, Tissue macrophage identity and self-renewal[J]. Immunol. Rev. 262(1), 56–73 (2014). https://doi.org/10.1111/imr.12224
S. Chakarov, H.Y. Lim, L. Tan et al., Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches[J]. Science 363(6432) (2019). https://doi.org/10.1126/science.aau0964
E. Evren, E. Ringqvist, K.P. Tripathi et al., Distinct developmental pathways from blood monocytes generate human lung macrophage diversity[J]. Immunity 54(2), 259–275 (2021). https://doi.org/10.1016/j.immuni.2020.12.003
R. Zilionis, C. Engblom, C. Pfirschke et al., Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species[J]. Immunity 50(5), 1317–1334 (2019). https://doi.org/10.1016/j.immuni.2019.03.009
Z. Chen, Y. Huang, Z. Hu et al., Landscape and dynamics of single tumor and immune cells in early and advanced-stage lung adenocarcinoma[J]. Clin. Transl. Med. 11(3), e350 (2021). https://doi.org/10.1002/ctm2.350
C. Hafemeister, R. Satija, Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression[J]. Genome Biol. 20(1), 296 (2019). https://doi.org/10.1186/s13059-019-1874-1
Z. Chen, Z. Hu, Q. Sui et al., LncRNA FAM83A-AS1 facilitates tumor proliferation and the migration via the HIF-1alpha/ glycolysis axis in lung adenocarcinoma[J]. Int. J. Biol. Sci. 18(2), 522–535 (2022). https://doi.org/10.7150/ijbs.67556
A.M. Newman, C.B. Steen, C.L. Liu et al., Determining cell type abundance and expression from bulk tissues with digital cytometry[J]. Nat. Biotechnol. 37(7), 773–782 (2019). https://doi.org/10.1038/s41587-019-0114-2
W.F. Hong, M.Y. Liu, L. Liang et al., Molecular characteristics of T Cell-mediated tumor killing in hepatocellular Carcinoma[J]. Front. Immunol. 13, 868480 (2022). https://doi.org/10.3389/fimmu.2022.868480
Z. Bian, Y. Gong, T. Huang et al., Deciphering human macrophage development at single-cell resolution[J]. Nature 582(7813), 571–576 (2020). https://doi.org/10.1038/s41586-020-2316-7
D. Lambrechts, E. Wauters, B. Boeckx et al., Phenotype molding of stromal cells in the lung tumor microenvironment[J]. Nat. Med. 24(8), 1277–1289 (2018). https://doi.org/10.1038/s41591-018-0096-5
E. Azizi, A.J. Carr, G. Plitas et al., Single-cell map of diverse immune phenotypes in the breast tumor microenvironment[J]. Cell 174(5), 1293–1308 (2018). https://doi.org/10.1016/j.cell.2018.05.060
A. Di Stefano, A. Capelli, M. Lusuardi et al., Severity of airflow limitation is associated with severity of airway inflammation in smokers[J]. Am. J. Respir. Crit. Care Med. 158(4), 1277–1285 (1998). https://doi.org/10.1164/ajrccm.158.4.9802078
S. Arandjelovic, K.S. Ravichandran, Phagocytosis of apoptotic cells in homeostasis[J]. Nat. Immunol. 16(9), 907–917 (2015). https://doi.org/10.1038/ni.3253
M. Liao, Y. Liu, J. Yuan et al., Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19[J]. Nat. Med. 26(6), 842–844 (2020). https://doi.org/10.1038/s41591-020-0901-9
X. Liang, K. Gupta, J.R. Quintero et al., Macrophage FABP4 is required for neutrophil recruitment and bacterial clearance in Pseudomonas aeruginosa pneumonia[J]. FASEB J. 33(3), 3562–3574 (2019). https://doi.org/10.1096/fj.201802002R
Y. Zhou, D. Yang, Q. Yang et al., Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma[J]. Nat. Commun. 11(1), 6322 (2020). https://doi.org/10.1038/s41467-020-20059-6
C. Sabatel, C. Radermecker, L. Fievez et al., Exposure to bacterial CpG DNA protects from airway allergic inflammation by expanding regulatory lung interstitial macrophages[J]. Immunity 46(3), 457–473 (2017). https://doi.org/10.1016/j.immuni.2017.02.016
J. Qi, H. Sun, Y. Zhang et al., Single-cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer[J]. Nat. Commun. 13(1), 1742 (2022). https://doi.org/10.1038/s41467-022-29366-6
P. Chen, D. Zhao, J. Li et al., Symbiotic macrophage-glioma cell interactions reveal synthetic lethality in PTEN-Null Glioma[J]. Cancer Cell 35(6), 868–884 (2019). https://doi.org/10.1016/j.ccell.2019.05.003
L. Zhang, Z. Li, K.M. Skrzypczynska et al., Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer[J]. Cell 181(2), 442–459 (2020). https://doi.org/10.1016/j.cell.2020.03.048
Q. Yang, H. Zhang, T. Wei et al., Single-Cell RNA sequencing reveals the heterogeneity of tumor-associated macrophage in non-small cell lung cancer and differences between sexes[J]. Front. Immunol. 12, 756722 (2021). https://doi.org/10.3389/fimmu.2021.756722
Y. Zhang, W. Du, Z. Chen et al., Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma[J]. Exp. Cell Res. 359(2), 449–457 (2017). https://doi.org/10.1016/j.yexcr.2017.08.028
A.M. Leader, J.A. Grout, B.B. Maier et al., Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification[J]. Cancer Cell 39(12), 1594–1609 (2021). https://doi.org/10.1016/j.ccell.2021.10.009
J. Chen, Y. Yao, C. Gong et al., CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3[J]. Cancer Cell 19(4), 541–555 (2011). https://doi.org/10.1016/j.ccr.2011.02.006
H. Ye, Q. Zhou, S. Zheng et al., Tumor-associated macrophages promote progression and the Warburg effect via CCL18/NF-kB/VCAM-1 pathway in pancreatic ductal adenocarcinoma[J]. Cell Death Dis. 9(5), 453 (2018). https://doi.org/10.1038/s41419-018-0486-0
Y. Wu, S. Yang, J. Ma et al., Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level[J]. Cancer Discov. 12(1), 134–153 (2022). https://doi.org/10.1158/2159-8290.CD-21-0316
H. Wang, B. Chen, Y. Lin et al., Legumain promotes gastric cancer progression through tumor-associated macrophages in vitro and In vivo[J]. Int. J. Biol. Sci. 16(1), 172–180 (2020). https://doi.org/10.7150/ijbs.36467
Q. Zhang, Y. He, N. Luo et al., Landscape and dynamics of single immune cells in hepatocellular carcinoma[J]. Cell 179(4), 829–845 (2019). https://doi.org/10.1016/j.cell.2019.10.003
C.M. Carnielli, C. Macedo, T. De Rossi et al., Combining discovery and targeted proteomics reveals a prognostic signature in oral cancer[J]. Nat. Commun. 9(1), 3598 (2018). https://doi.org/10.1038/s41467-018-05696-2
X. Yuan, Q. Zhao, Y. Zhang et al., The role and mechanism of HLA complex group 11 in cancer[J]. Biomed. Pharmacother. 143, 112210 (2021). https://doi.org/10.1016/j.biopha.2021.112210
R. Pio, D. Ajona, S. Ortiz-Espinosa et al., Complementing the cancer-immunity cycle[J]. Front. Immunol. 10, 774 (2019). https://doi.org/10.3389/fimmu.2019.00774
A. Alquraini, K.J. El, Scavenger receptors[J]. Curr. Biol. 30(14), R790–R795 (2020). https://doi.org/10.1016/j.cub.2020.05.051
D. Chowell, L. Morris, C.M. Grigg et al., Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy[J]. Science 359(6375), 582–587 (2018). https://doi.org/10.1126/science.aao4572
M.B. Buechler, W. Fu, S.J. Turley, Fibroblast-macrophage reciprocal interactions in health, fibrosis, and cancer[J]. Immunity 54(5), 903–915 (2021). https://doi.org/10.1016/j.immuni.2021.04.021
Y. Zhu, J. Yang, D. Xu et al., Disruption of tumour-associated macrophage trafficking by the osteopontin-induced colony-stimulating factor-1 signalling sensitises hepatocellular carcinoma to anti-PD-L1 blockade[J]. Gut 68(9), 1653–1666 (2019). https://doi.org/10.1136/gutjnl-2019-318419
V. Quaranta, C. Rainer, S.R. Nielsen et al., Macrophage-derived granulin drives resistance to immune checkpoint inhibition in metastatic pancreatic cancer[J]. Cancer Res. 78(15), 4253–4269 (2018). https://doi.org/10.1158/0008-5472.CAN-17-3876
X. Yang, Y. Lin, Y. Shi et al., FAP promotes immunosuppression by cancer-associated fibroblasts in the tumor microenvironment via STAT3-CCL2 signaling[J]. Cancer Res. 76(14), 4124–4135 (2016). https://doi.org/10.1158/0008-5472.CAN-15-2973
M. Augsten, C. Hagglof, E. Olsson et al., CXCL14 is an autocrine growth factor for fibroblasts and acts as a multi-modal stimulator of prostate tumor growth[J]. Proc. Natl. Acad. Sci. U. S. A. 106(9), 3414–3419 (2009). https://doi.org/10.1073/pnas.0813144106
R. Allaoui, C. Bergenfelz, S. Mohlin et al., Cancer-associated fibroblast-secreted CXCL16 attracts monocytes to promote stroma activation in triple-negative breast cancers[J]. Nat. Commun. 7, 13050 (2016). https://doi.org/10.1038/ncomms13050
G. Ren, X. Zhao, Y. Wang et al., CCR2-dependent recruitment of macrophages by tumor-educated mesenchymal stromal cells promotes tumor development and is mimicked by TNFalpha[J]. Cell Stem Cell 11(6), 812–824 (2012). https://doi.org/10.1016/j.stem.2012.08.013
I. Martinez-Reyes, N.S. Chandel, Cancer metabolism: looking forward[J]. Nat. Rev. Cancer 21(10), 669–680 (2021). https://doi.org/10.1038/s41568-021-00378-6
O.R. Colegio, N.Q. Chu, A.L. Szabo et al., Functional polarization of tumour-associated macrophages by tumour-derived lactic acid[J]. Nature 513(7519), 559–563 (2014). https://doi.org/10.1038/nature13490
Y. Sun, Z. Zheng, H. Zhang et al., Chemotherapeutic tumor microparticles combining low-dose irradiation reprogram tumor-promoting macrophages through a tumor-repopulating cell-curtailing pathway[J]. Oncoimmunology 6(6), e1309487 (2017). https://doi.org/10.1080/2162402X.2017.1309487
J.Y. Wu, T.W. Huang, Y.T. Hsieh et al., Cancer-derived succinate promotes macrophage polarization and cancer metastasis via succinate receptor[J]. Mol. Cell 77(2), 213–227 (2020). https://doi.org/10.1016/j.molcel.2019.10.023
B. Allard, P.A. Beavis, P.K. Darcy et al., Immunosuppressive activities of adenosine in cancer[J]. Curr. Opin. Pharmacol. 29, 7–16 (2016). https://doi.org/10.1016/j.coph.2016.04.001
F. Martinez-Jimenez, F. Muinos, I. Sentis et al., A compendium of mutational cancer driver genes[J]. Nat. Rev. Cancer 20(10), 555–572 (2020). https://doi.org/10.1038/s41568-020-0290-x
S. Hodge, G. Hodge, J. Ahern et al., Smoking alters alveolar macrophage recognition and phagocytic ability: implications in chronic obstructive pulmonary disease[J]. Am. J. Respir. Cell Mol. Biol 37(6), 748–755 (2007). https://doi.org/10.1165/rcmb.2007-0025OC
S.T. Lugg, A. Scott, D. Parekh et al., Cigarette smoke exposure and alveolar macrophages: mechanisms for lung disease[J]. Thorax 77(1), 94–101 (2022). https://doi.org/10.1136/thoraxjnl-2020-216296
S.G. Conticello, The AID/APOBEC family of nucleic acid mutators[J]. Genome Biol. 9(6), 229 (2008). https://doi.org/10.1186/gb-2008-9-6-229
Acknowledgements
We thank Shanghai OE Biotech Company (Shanghai, China) for carrying out the scRNA-Seq analysis.
Funding
This article is sponsored by Shanghai Sailing Program (22YF1407500).
Author information
Authors and Affiliations
Contributions
Made a substantial contribution to the concept or design of the work: Zhengyang Hu, Xing Jin, Weifeng Hong, Qihai Sui, Mengnan Zhao, Yiwei Huang, Ming Li, Qun Wang, Cheng Zhan, Zhencong Chen
Acquisition, analysis or interpretation of data: Zhengyang Hu, Xing Jin, Weifeng Hong, Qihai Sui, Mengnan Zhao, Zhencong Chen
Drafted the article or revised it critically for important intellectual content: Zhengyang Hu, Xing Jin, Yiwei Huang, Ming Li, Qun Wang, Cheng Zhan, Zhencong Chen
Approved the version to be published: Zhengyang Hu, Xing Jin, Weifeng Hong, Qihai Sui, Mengnan Zhao, Yiwei Huang, Ming Li, Qun Wang, Cheng Zhan, Zhencong Chen
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was approved by the Ethics Committee of Zhongshan Hospital, Fudan University, China (B2018–137R). The informed consent was obtained at the time of hospitalization.
Consent for publication
We have obtained written informed consent from the patient and the written consent is in the patient's hospital record.
Conflicts of interest
The authors declare no competing interests in this work.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, Z., Jin, X., Hong, W. et al. Dissecting the single-cell transcriptome network of macrophage and identifies a signature to predict prognosis in lung adenocarcinoma. Cell Oncol. 46, 1351–1368 (2023). https://doi.org/10.1007/s13402-023-00816-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-023-00816-7