Abstract
Objectives
Previously, Interferon-induced Protein with Tetratricopeptide Repeats 1 (IFIT1) has been shown to promote cancer development. Here, we aimed to explore the role of IFIT1 in the development and progression of pancreatic cancer, including the underlying mechanisms.
Methods
We explored IFIT1 expression in pancreatic cancer samples using The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets. Cell Counting Kit-8 (CCK8), colony formation, scratch wound-healing and Transwell assays were performed to assess the proliferation, migration and invasion abilities of pancreatic cancer cells. Gene Set Enrichment Analysis (GSEA) and Western blotting were performed to assess the regulatory effect of IFIT1 on the Wnt/β-catenin pathway.
Results
We found that upregulation of IFIT1 expression is common in pancreatic cancer and is negatively associated with overall patient survival. Knockdown of IFIT1 expression led to decreased proliferation, migration and invasion of pancreatic cancer cells. We also found that IFIT1 could regulate Wnt/β-catenin signaling, and that a Wnt/β-catenin agonist could reverse this effect. In addition, we found that IFIT1 can promote epithelial-mesenchymal transition (EMT) of pancreatic cancer cells.
Conclusions
Our data indicate that IFIT1 increases pancreatic cancer cell proliferation, migration and invasion by activating the Wnt/β-catenin pathway. In addition, we found that EMT could be regulated by IFIT1. IFIT1 may serve as a potential therapeutic target for pancreatic cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
As the fourth leading cause of tumor-related death, pancreatic ductal adenocarcinoma (PDAC) is the most malignant cancer in the world, with a 5-year overall survival (OS) rate lower than 7% [1, 2]. At present, surgical resection is the only potential cure. However, patients with PDAC are usually diagnosed at a late stage with locally advanced or metastatic disease [3]. In addition, most patients who undergo resection experience recurrence and metastasis [4]. Although several therapies and technologies have been used to treat and diagnose PDAC patients, the mortality rate continues to increase by 0.3% each year [5]. There are some reasons that could explain this trend. First, because of the limitations of detection technologies and specific markers, pancreatic cancer patients are usually diagnosed at advanced stages. Second, pancreatic cancer is often resistant to radiotherapy, chemotherapy, immune therapy and molecular targeted therapy. In addition, complex molecular and cellular changes in pancreatic cancer pose a substantial challenge for treatment efficiency [6, 7].
Epithelial-mesenchymal transition (EMT) is a complex process by which epithelial cells acquire mesenchymal traits and lose their cell-cell junctions. EMT is characterized by upregulation of N-cadherin, fibronectin and vimentin expression and downregulation of E-cadherin expression [8, 9]. In pancreatic cancer it has been shown that EMT plays a significant role in the processes of tumor progression and metastasis [10,11,12]. Several pathways have been correlated with the modulation of Wnt/β-catenin signaling, with EMT being one of the most canonical ones [13, 14]. Wnt/β-catenin signaling can effectively influence the progression of pancreatic cancer [15]. Once Wnt ligands bind to their family member receptors, this pathway can be activated by the nuclear transport of β-catenin. By subsequently binding to TCF/LEF transcription factors, β-catenin can activate the transcription of a series of EMT-related marker genes in the nucleus [16, 17].
Modulated by the JAK/STAT pathway, IFIT1 is known as an inflammation-related protein that can be aberrantly expressed in cancer [18]. As RNA-binding proteins, the IFIT protein family has been found to inhibit the translation of non-self RNA in the process of viral infection [19]. Its tetratricopeptide repeat (TPR) motifs have been shown to regulate the homodimerization of IFIT protein family members [19]. Although most attention has focused on the correlation of IFITs with antiviral characteristics, recent evidence indicates that the IFIT protein family may also play an important role in cancer development [20, 21]. Liu et al. suggested that IFIT1 and IFIT3 expression could modulate cell migration and metastasis in patients with hepatocellular carcinoma [21]. Pidugu et al. showed that IFIT1/3 could regulate resistance to cisplatin and modulate the metastatic ability of oral squamous cell carcinoma (OSCC) cells [20]. In addition, IFIT1/3 has been shown to enhance sensitivity to gefitinib by increasing p-EGFR recycling in OSCC cells [20]. At present, the exact role of IFIT1 in pancreatic cancer development is still unknown.
First, we investigated IFIT1 expression in PDAC tissues by analyzing TCGA and GEO datasets and by performing immunohistochemistry. Next, we evaluated the relationship of IFIT1 expression with patient survival and clinicopathological features. In addition, we evaluated the role of IFIT1 in the proliferation, invasion and migration of pancreatic cancer cells using functional assays. GSEA revealed that MMP cytokine-, TGF-β- and Wnt/β-catenin-associated gene sets were enriched in pancreatic cancer samples with high IFIT1 expression levels. In addition, we found that IFIT1 could regulate the expression of Wnt/β-catenin pathway-related markers and EMT markers. A Wnt/β-catenin signaling pathway activator rescued the effects of IFIT1 knockdown on pancreatic cancer cell proliferation and migration. IFIT1 may serve as a potential target for the treatment of pancreatic cancer patients.
2 Materials and methods
2.1 Bioinformatics analysis
mRNA expression data were acquired from TCGA and GEO datasets. Survival data of the pancreatic cancer samples were analyzed using a log-rank test. Samples from TCGA datasets with a high IFIT1 expression (above the mean) were denoted as a group with high IFIT1 expression, and samples with a low IFIT1 expression (below the mean) were denoted as a group with low IFIT1 expression. To explore crucial biological pathways modulated by IFIT1, GSEA was performed based on expression data from patients with low and high expression levels.
2.2 Tissue microarray analysis
A tissue microarray was acquired from Zuocheng Biotech (Shanghai, China) containing 76 pancreatic tumor samples and 76 normal adjacent pancreatic samples. The patients enrolled in our study received little preoperative treatment. The tissues were incubated with a primary anti-IFIT1 antibody (1:500 dilution), after which the staining results were evaluated by microscopy (Nikon ECLIPSETs2R). The results were scored by two pathologists independently as follows: cells stained less than 10% were rated as negative (−, 1), cells stained between 10 and 49% were rated as (+, 2), cells stained between 50 and 74% were rated as (++, 3) and cells stained between 75 and 100% were rated as (+++, 4). The intensity/color of staining was rated as negative particles, lightly yellow particles [22], brownish yellow particles [23] and brown particles [24]. We defined the final scores by multiplying the staining number scores by the staining color scores [25]. A score between 0 and 5 was defined as negative and a score > 5 as positive expression.
2.3 Cell lines and culture conditions
The pancreatic cancer cell lines AsPC-1, MiaPaCa-2, BxPC-3, Patu8988, Panc-1 and CFPAC-1 were obtained from the American Type Culture Collection (ATCC). The cell lines were seeded in DMEM, RPMI-1640 or IMDM (HyClone) media supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% antibiotics and grown in a cell incubator at 37 °C with 5% CO2.
2.4 Transfection assay
Cells were seeded and cultured in 6-well plates at 37 °C for 24 h. When the cells reached 50–70% confluence, they were infected with an IFIT1-overexpression plasmid (IFIT1- OE) or a negative control plasmid (IFIT1-NC) (Tsingke, China). IFIT1 cDNA was synthesized and subcloned into a pcDNA3.1(+) vector using its BamHI and XhoI restriction sites. A small interfering IFIT1 RNA (si-IFIT1) (siRNA: 5’-CCUUGGAAUACUACACUCA-3′) and a negative control siRNA (siNC) (siRNA: 5′- UUCUCCGAACGUGUCACGU-3′) were constructed by RiboBio (Guangzhou, China). Lipofectamine 3000 (Invitrogen, USA) was used according to the manufacturer’s instructions. The efficiency of the transfections was investigated after 48–72 h using Western blotting (WB) and RT-PCR.
2.5 Western blotting
Western blotting assays were performed as described previously [26]. The primary antibodies used were: anti-IFIT1 (Proteintech, 23,247–1-AP), anti-E-cadherin (Proteintech, 20,874–1-AP), anti-N-cadherin (Proteintech, 22,018–1-AP), anti-Vimentin (Proteintech, 10,366–1-AP), anti-MMP9 (CST, 2270), anti-β-catenin (CST, 8480), anti-Cyclin D1 (Abcam, AB16663), anti-c-Myc (CST, 5605), anti-survivin (CST, 2808) and anti-GAPDH (Proteintech, 10,494–1-AP).
2.6 RNA isolation and RT-PCR
RNA was extracted from treated pancreatic cancer cells using TRIzol reagent (Ambion, Life Technologies). cDNA was generated using a reverse transcription kit (Takara, China) in a 96-well thermal cycler (Applied Biosystems). Quantitative PCR was conducted using SYBR Premix Ex TaqTM-Reagent (TakaRa, China) by applying StepOnePlus™ (Applied Biosystems) based on the manufacturer’s instructions. The IFIT1 forward primer used was TGAGCCTCCTTGGGTTCGTCTAC, and the IFIT1 reverse primer used was CTCAAAGTCAGCAGCCAGTCTCAG. β-actin forward: CTCCATCCTGGCCTCGCT GT; β-actin reverse: GCTGTCACCTTCACCGTTCC. Fold changes in expression relative to β-actin were calculated using the 2–DDCt method.
2.7 Cell viability and colony formation assays
Pancreatic cancer cells were seeded in 96-well plates (2.5 × 103 cells per well) after which viability was measured at 5 time points (0 h, 24 h, 48 h, 72 h and 96 h) using a CCK8 assay (Dojindo) at 450 nm. To investigate proliferation, we performed a colony formation assay. After preparing single cell suspensions, 1200 cells were seeded in each well of a 6-well plate. After 11–14 days, the cells were fixed in 3.7% formaldehyde (Sigma-Aldrich) and stained with 0.4% crystal violet (Sigma-Aldrich). Each plate was washed by 3 thorough immersions in pure water and then scanned.
2.8 Transwell migration and invasion assays
The upper chambers of a Transwell plate (24-well, 8-μm, Corning, Life Sciences) were coated with Matrigel (BD Biosciences, USA) for the invasion assay, whereas no Matrigel was used for the migration assay. Approximately 3 × 104 cells were seeded in the upper chamber and incubated overnight. Culture medium with 10% FBS was placed in the lower chamber, and culture medium without FBS was placed in the upper chamber. After incubation at 37 °C for 48 h the nonmigrating cells were gently removed. The migrated/invaded cells were fixed in 90% ethyl alcohol and then stained with 1% crystal violet. Cells from 5 random fields were counted under an inverted microscope.
2.9 Statistical analysis
The results are presented as mean ± standard deviation. Categorical variables were analyzed by applying the chi-square test, and continuous variables were analyzed by applying the t-test. Fisher’s test was applied to explore associations between IFIT1 expression and clinicopathological features. Kaplan-Meier analysis and the log-rank test were applied for overall survival analysis. Multivariate prognostic predictors were calculated by Cox regression analysis. Statistical analyses were performed using SPSS 23.0 software (SPSS, Chicago, USA) and GraphPad Prism 7.0 (La Jolla, USA). A p value < 0.05 was considered statistically significant.
3 Results
3.1 The expression level of IFIT1 is markedly higher in pancreatic cancer tissues than in normal tissues
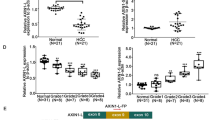
To determine IFIT1 expression in pancreatic cancer, we referred to the TCGA, GEO (GSE16515, GSE15471) and Genotype-Tissue Expression (GTEx) databases and extracted information from the expression profiles of the datasets. We found that the expression level of IFIT1 in the pancreatic cancer tissues was significantly increased compared to that in the normal tissues in all these datasets (Fig. 1A-C). To substantiate the putative role of IFIT1 in pancreatic cancer, we measured IFIT1 protein expression in 76 pancreatic cancer samples and paired normal adjacent tissues using immunohistochemistry (IHC). By comparing the H-scores, we found that the protein levels of IFIT1 were significantly increased in pancreatic cancer samples (p = 0.01; Fig. 1D-F). By exploring the association between IFIT1 expression and clinicopathologic variables, we found that the expression of IFIT1 was significantly related to pathological stage (p = 0.002) and tumor size (p = 0.042; Table 1). In addition, we explored associations between clinicopathological features and the expression of IFIT1 using the TCGA dataset. We observed significant associations between IFIT1 expression and N stage (p = 0.047), histological grade (p < 0.001) and smoking history (p = 0.026; Supplementary Table 1).
Expression of IFIT1 in pancreatic ductal adenocarcinoma tissues. (A-C) Differential expression levels of IFIT1 in pancreatic cancer. (D-F) Expression of IFIT1 in PDAC tissues and peritumor tissues as detected by IHC. (G, H) High expression of IFIT1 correlates with a poor prognosis in pancreatic cancer patients
3.2 IFIT1 expression is an independent prognostic factor for pancreatic cancer patients
Next, we assessed whether IFIT1 may be used as a prognostic predictor in pancreatic cancer patients. Kaplan-Meier (KM) results from the TCGA database indicated that a higher IFIT1 expression was significantly associated with a poorer survival (p = 0.039; Fig. 1G). Additionally, we analyzed a putative correlation between the expression level of IFIT1 and clinical follow-up data in 76 pancreatic cancer patient samples. We found that the patients in the higher expression group showed a markedly poorer overall survival than patients in the lower expression group (p < 0.001; Fig. 1H). Additionally, univariate and multivariate Cox analyses were performed to explore the association between the IFIT1 IHC data and clinicopathological features (Table 2). We found that the following factors were significantly related to OS: T stage (hazard ratio [HR] 1.900; 95% CI 1.098–3.288; p = 0.022), sex (HR 2.399; 95% CI 1.288–4.468; p = 0.006), tumor size (HR 1.744; 95% CI 1.021–2.980; p = 0.042) and cytoplasmic expression of IFIT1 (HR 3.154; 95% CI 1.763–5.642; p < 0.001). In multivariate Cox analysis models, male sex (HR 2.992; 95% CI 1.549–5.779; p < 0.001), T stage III-IV (HR 1.923; 95% CI 0.993–3.722; p = 0.052) and higher expression of IFIT1 (HR 4.018; 95% CI 2.160–7.475; p < 0.001) were correlated with OS (Table 2). Therefore, we conclude that IFIT1 expression may serve as an independent biomarker for the prognosis of pancreatic cancer patients.
3.3 IFIT1 expression in pancreatic cancer cell lines
Using RT-PCR and WB we found that IFIT1 was expressed in all 6 pancreatic cancer cell lines tested (Fig. 2A, B and Fig. S1A). BxPC-3 cells exhibited a relatively higher expression level, whereas PANC-1 cells exhibited a relatively lower expression level. AsPC-1, MiaPaCa-2, Patu-8988 and CFPAC-1 cells exhibited moderate IFIT1 expression levels (Fig. 2A, B and Fig. S1A). To explore a putative biological function of IFIT1 in PDAC, we overexpressed IFIT1 in AsPC-1 and PANC-1 cells by transfection with an IFIT1 OE plasmid and downregulated IFIT1 expression in AsPC-1 and Bxpc-3 cells by transfection with an IFIT1 short interference RNA (siRNA). The efficacy of IFIT1-OE and si-IFIT1 compared to the negative controls was verified by RT-PCR and Western blotting (Fig. 2C, D; Fig. S1B, C).
IFIT1 expression in pancreatic cancer cell lines and transient transduction of IFIT1 overexpression and knockdown constructs. (A, B) Protein level of IFIT1 in 6 pancreatic cancer cell lines. (C) Efficiency of IFIT1 knockdown in Aspc-1 and Bxpc-3 cells confirmed by Western blotting. (D) Efficiency of IFIT1 overexpression in Aspc-1 and Panc-1 cells confirmed by Western blotting. GAPDH was used as internal control
3.4 IFIT1 increases the proliferation of pancreatic cancer cells
To investigate the function of IFIT1 in the development of pancreatic cancer, CCK8 and colony formation assays were performed using pancreatic cancer cell lines. The viability of the cells assessed by the CCK8 assay indicated that IFIT1 expression knockdown reduced the growth of BxPC-3 and AsPC-1 cells (p < 0.05; Fig. 3A, B). The colony formation assay results revealed that IFIT1 expression knockdown decreased the colony numbers by approximately 60% compared to the controls (p < 0.05; Fig. 3E). These results indicate that IFIT1 may play an important role in modulating the proliferative ability of pancreatic cancer cells. To further explore the influence of IFIT1 on the proliferation of pancreatic cancer cells, an IFIT1 OE plasmid was constructed and transfected into AsPC-1 and PANC-1 cells. Consistent with the results of IFIT1 knockdown, we found that IFIT1 OE promoted the viability of AsPC-1 and PANC-1 cells (p < 0.05; Fig. 3C, D). In addition, we found that the colony forming capacity of AsPC-1 and PANC-1 cells transfected with IFIT1 OE was nearly 1.6-fold higher than that of the control cells (p < 0.05; Fig. 3F). Overall, these analyses indicate that IFIT1 may act as an important factor regulating the proliferation of pancreatic cancer cells.
IFIT1 enhances the proliferation of pancreatic cancer cells. A CCK8 assay was performed to determine the proliferation of IFIT1 knockdown (A, B) or overexpressing (C, D) pancreatic cancer cells at the indicated time points after seeding. Cell viability was measured at 450 nm. The effect of IFIT1 knockdown (E) or overexpression (F) on the colony forming capacity of pancreatic cancer cells is shown in the left panels. The number of foci was counted as shown in the right panels. All the data are presented as mean ± SEM
3.5 IFIT1 enhances the invasion and migration of pancreatic cancer cells
To analyze the role of IFIT1 in the invasion and migration of pancreatic cancer cells, we performed Transwell and scratch wound-healing assays. We found that in comparison with the control cells, the IFIT1 KD BxPC-3 and AsPC-1 cells exhibited prominent decreases in their wound healing abilities (p < 0.05; Fig. 4A, B). Furthermore, using a Transwell assay, we found that the migration rate of the IFIT1 KD cells was lower than that of the control cells (p < 0.01; Fig. 5A). In comparison with the IFIT1 KD group, the migration capacity of AsPC-1 and PANC-1 cells was significantly increased in the IFIT1 OE group (p < 0.01; Fig. 4C, D and Fig. 5B). We also analyzed the effect of IFIT1 expression on the metastatic capacity of pancreatic cancer cells by conducting Transwell invasion experiments. We found that in comparison with the controls, IFIT1 KD significantly decreased the invasive ability of Bxpc-3 and AsPC-1 cells (p < 0.01; Fig. 5C), while IFIT1 OE increased the invasive ability of these cells (p < 0.01; Fig. 5D). Based on these results, we conclude that IFIT1 may enhance the migrative and invasive abilities of pancreatic cancer cells.
IFIT1 promotes the migration ability of PC cells. (A-D) Effect of IFIT1 knockdown (A, B) or IFIT1 overexpression (C, D) on pancreatic cancer cell migration detected by wound-healing assays at the indicated time points after scratching. Wound healing was measured using ImageJ software. The data are presented as mean ± SEM. *p < 0.05, ** p < 0.01, *** p < 0.001
IFIT1 promotes the migration and invasion and induces EMT in pancreatic cancer cells. (A, B) Motility of IFIT1 knockdown (A) or IFIT1 overexpressing (B) pancreatic cancer cells assessed by Transwell assay at 24 h. Representative images of migration were captured at 24 h (left panels). (C, D) Invasive ability of IFIT1 knockdown (C) or IFIT1 overexpressing (D) pancreatic cancer cells assessed by Transwell assay at 48 h. Representative images of invasion were captured at 48 h (left panels). The data are presented as mean ± SEM. (E) Morphology of IFIT1 overexpressing and control AsPC-1 and Panc-1 cells. (F, G) Protein levels of EMT-related markers in IFIT1-knockdown (F) or overexpressing (G) cells assessed by Western blotting. GAPDH was used as internal control. * p < 0.05, ** p < 0.01, *** p < 0.001
3.6 IFIT1 regulates EMT in pancreatic cancer cells
EMT is closely associated with cell motility [8]. Therefore, we next analyzed whether EMT may be influenced by IFIT1-mediated changes in the invasive and migrative abilities of pancreatic cancer cells. We found that both AsPC-1 and Panc-1 cells lost their epithelial cell characteristics and, instead acquired fibroblast-like spindle-shaped morphologies (Fig. 5E). In addition, we explored the expression levels of E-cadherin, N-cadherin and Vimentin by performing RT-PCR and Western blotting. We found that in IFIT1 KD cells the expression of E-cadherin, a conventional epithelial marker, was significantly upregulated, and that the expression of N-cadherin and Vimentin, known as mesenchymal markers, was downregulated significantly (Fig. 5F, G). Furthermore, we found that the expression of a member of the matrix metalloproteinase family, MMP9, which is associated with migration and invasion, was significantly decreased in the IFIT1 KD cells (Fig. 5F). Conversely, we found that IFIT1 OE led to an increased E-cadherin expression and a decreased N-cadherin, Vimentin and MMP9 expression (Fig. 5G). These results suggest a role of IFIT1 in the EMT of pancreatic cancer cells.
3.7 GSEA indicates that IFIT1 expression correlates with EMT-related gene expression
To explore the mechanisms by which IFIT1 promotes the malignant behavior of pancreatic cancer cells we conducted GSEA. We found that IFIT1 expression correlates with pancreatic cancer (Fig. 6A). In addition, TGF-β and MMP cytokine connection gene sets were found to be enriched in the IFIT1 high expression group (Fig. 6B, C). Also, the β-catenin-associated gene set was found to be significantly affected by different IFIT1 expression levels (Fig. 6D).
IFIT1-induced EMT is regulated by the Wnt/beta-catenin signaling pathway. (A-D) GSEA revealing pancreatic cancer-, TGF-β signaling-, MMP cytokine- and Wnt/β-catenin-related gene sets whose expression was related to IFIT1 expression. (E, F) β-catenin, cyclin D1, c-Myc and survivin protein levels assayed by Western blotting in IFIT1 knockdown (E) or IFIT1 overexpressing (F) pancreatic cancer cells. GAPDH was used as internal control
3.8 IFIT1 modulates EMT by regulating Wnt/β-catenin signaling
β-catenin is a key factor in the Wnt/β-catenin signaling pathway and plays an important role in the progression of pancreatic cancer [27]. In several studies it has been found that a high expression of β-catenin may induce EMT [28]. To explore the mechanism by which IFIT1 regulates EMT in pancreatic cancer cells, we explored β-catenin expression and downstream signaling molecules of the Wnt/β-catenin pathway, including c-Myc, cyclin D1 and survivin [28, 29]. In the IFIT1 KD cells, β-catenin expression was found to be decreased (Fig. 6E). In contrast, we found that IFIT1 OE resulted in increased β-catenin expression at both the protein and RNA levels, which indicates that IFIT1 may regulate the expression of β-catenin at the transcriptional level (Fig. 6F; Supplementary Fig. 1D, E). Next, through Western blotting, we examined the expression level of the downstream molecules of the Wnt/β-catenin pathway. In the IFIT1 KD cells, the expression of cyclin D1, c-Myc and survivin in pancreatic cancer cells was found to be significantly decreased (Fig. 6E). Conversely, we found that IFIT1 OE increased the expression level of the above genes (Fig. 6F). To further explore the association between Wnt/β-catenin signaling and IFIT1 knockdown, AsPC-1 and BxPC-3 cells were cultured in the presence of 6 μM CHIR99021, a specific activator of the Wnt/β-catenin pathway [29]. A subsequent CCK8 assay revealed that CHIR99021 significantly increased the proliferation rate of the IFIT1-KD AsPC-1 and BxPC-3 cells (Fig. 7A, B), as well as their colony forming ability (Fig. 7C, D). In addition, we found that the migration rate of the IFIT1-KD AsPC-1 and BxPC-3 cells was increased by CHIR99021 through activation of the Wnt/β-catenin pathway (Fig. 7E, F). CHIR99021 rescued the reduction in gene expression caused by IFIT1 KD (Fig. 7G). These results indicate that IFIT1 expression leads to pancreatic cancer progression in a Wnt/β-catenin-dependent manner.
IFIT1 regulates pancreatic cancer cell proliferation and migration through the β-catenin pathway. (A-D) Scramble and IFIT1-KD pancreatic cancer cells were pretreated with DMSO or CHIR99021 (6 μM/ml) for 24 h, after which cell proliferation was determined by (A, B) growth curve and (C, D) colony formation assays. (E, F) Cell migration determined by wound healing assay in pretreated Aspc-1 cells. (G) Protein levels of EMT markers and Wnt/β-catenin target genes (β-catenin, cyclin D1, c-Myc and survivin) detected by Western blotting. GAPDH was used as internal control. * p < 0.05, ** p < 0.01, *** p < 0.001
4 Discussion
The IFIT family of genes, also known as interferon-stimulating genes, has extensively been studied mainly due to its antiviral properties [30]. All IFITs share similar motifs, comprising a helix-turn-helix structure that can form multiple types of protein complexes [31, 32]. Additional recent studies have indicated that IFITs may act as prognostic biomarkers for clinicopathological outcomes of multiple cancers, including breast cancer, hepatocellular carcinoma, pancreatic cancer and glioblastoma [33,34,35,36]. Previous studies have also indicated that IFIT1 may act as a tumor promotor and that its expression is elevated in several cancer types. After examining the GEO and TCGA datasets and primary pancreatic cancer tissues, we found that the expression of IFIT1 was higher in pancreatic tumor tissues than in adjacent normal tissues. Furthermore, we found that a higher expression of IFIT1 was positively associated with pathological stage and tumor size and that a higher IFIT1 expression was correlated with a worse survival. In addition, we found that IFIT1 expression knockdown resulted in a decreased proliferation, migration and invasion of pancreatic cancer cells.
EMT is a typical process that occurs in various tumors during which cells with epithelial features escape structural restraints by changing their cell type and cytoskeleton. Activation of EMT is considered to be a cause of the metastasis cascade [37,38,39,40]. Andrew et al. reported that the metastasis of pancreatic cancer cells is correlated with EMT [12]. We found that a decrease in IFIT1 expression increased the expression of E-cadherin and decreased the expression of N-cadherin, Vimentin and MMP9. Conversely, we found that IFIT1 OE increased the expression of N-cadherin, Vimentin and MMP9 while decreasing the expression of E-cadherin. These results indicated that IFIT1 may induce EMT in pancreatic cancer cells.
Next, we performed GSEA to identify IFIT1 downstream signaling pathways and gene sets correlated with IFIT1 expression. We found that in pancreatic cancer TGF-β receptor signaling, MMP cytokine production and the Wnt/β-catenin pathway were significantly related to IFIT1 expression. Among these, the Wnt/β-catenin signaling pathway has been reported to have a major effect on EMT [29]. We also found that the expression of β-catenin, c-Myc, cyclin D1 and survivin were upregulated or downregulated by IFIT1 KD or OE, respectively. In addition, we found that the inhibitory effect of IFIT1 KD on the biological behavior of AsPC-1 and BxPC-3 cells and the changes in β-catenin and downstream target gene expression modulated by IFIT1 KD could be rescued by treatment with CHIR99021, an activator of the Wnt/β-catenin pathway. These results indicate that IFIT1 can modulate the Wnt/β-catenin pathway in pancreatic cancer cells.
In conclusion, we found that IFIT1 may promote the proliferation, migration and invasion of pancreatic cancer cells, which is regulated by Wnt/β-catenin signaling. Additionally, we found that IFIT1 may facilitate EMT in pancreatic cancer. These results indicate that a combination of IFIT1 and Wnt/β-catenin pathway suppression may be an effective novel method for pancreatic cancer treatment. Limitations still exist in this work. First, we did not perform in vivo proliferation and/or invasion assays. Second, although we showed that IFIT1 may promote the malignant phenotype of pancreatic cancer cells through the Wnt/β-catenin pathway, direct evidence showing how IFIT1 interacts with the Wnt/β-catenin pathway is lacking. Hence, further research is warranted to substantiate the correlation between IFIT1 and the Wnt/β-catenin pathway in pancreatic cancer.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. The data are not publicly available due to privacy or ethical restrictions.
Change history
30 October 2023
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s13402-023-00897-4
Abbreviations
- ATCC:
-
American Type Culture Collection
- CCK8:
-
Cell Counting Kit-8
- EMT:
-
Epithelial-Mesenchymal Transition
- FBS:
-
Fetal Bovine Serum
- GEO:
-
Gene Expression Omnibus
- GSEA:
-
Gene Set Enrichment Analysis
- GTEx:
-
Genotype-Tissue Expression
- HR:
-
Hazard Ratio
- IFIT1:
-
Interferon-induced Protein with Tetratricopeptide Repeats 1
- IHC:
-
Immunohistochemistry
- KD:
-
Knockdown
- KM:
-
Kaplan-Meier
- OE:
-
Overexpression
- OS :
-
Overall Survival
- OSCC:
-
Oral Squamous Cell Carcinoma
- PDAC:
-
Pancreatic Ductal Adenocarcinoma
- RT-PCR:
-
Reverse Transcription-Polymerase Chain Reaction
- TRP:
-
Tetratricopeptide Repeat
- TCGA:
-
The Cancer Genome Atlas
References
T. Kamisawa, L.D. Wood, T. Itoi, K. Takaori, Pancreatic cancer. Lancet 388, 73–85 (2016)
C. Neuzillet, A. Tijeras-Raballand, P. Bourget, J. Cros, A. Couvelard, A. Sauvanet, M.-P. Vullierme, C. Tournigand, P. Hammel, State of the art and future directions of pancreatic ductal adenocarcinoma therapy. Pharmacol. Ther. 155, 80–104 (2015)
S. Gillen, T. Schuster, C.M.Z. Büschenfelde, H. Friess, J. Kleeff, Preoperative/neoadjuvant therapy in pancreatic cancer: A systematic review and meta-analysis of response and resection percentages. PLoS Med. 7, e1000267 (2010)
V.P. Groot, N. Rezaee, W. Wu, J.L. Cameron, E.K. Fishman, R.H. Hruban, M.J. Weiss, L. Zheng, C.L. Wolfgang, J. He, Patterns, timing, and predictors of recurrence following pancreatectomy for pancreatic ductal adenocarcinoma. Ann. Surg. 267, 936–945 (2018)
R.L. Siegel, K.D. Miller, A. Jemal, Cancer statistics, 2017. CA Cancer J. Clin. 67, 7–30 (2017)
J. He, N. Ahuja, M.A. Makary, J.L. Cameron, F.E. Eckhauser, M.A. Choti, R.H. Hruban, T.M. Pawlik, C.L. Wolfgang, 2564 resected periampullary adenocarcinomas at a single institution: Trends over three decades. HPB (Oxford) 16, 83–90 (2014)
D. Hanahan, R.A. Weinberg, Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011)
H. Acloque, M.S. Adams, K. Fishwick, M. Bronner-Fraser, M.A. Nieto, Epithelial-mesenchymal transitions: The importance of changing cell state in development and disease. J. Clin. Invest. 119, 1438–1449 (2009)
R. Kalluri, R.A. Weinberg, The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 (2009)
S. Wang, S. Huang, Y.L. Sun, Epithelial-mesenchymal transition in pancreatic Cancer: A review. Biomed. Res. Int. 2017, 2646148 (2017)
N. Gaianigo, D. Melisi, C. Carbone, EMT and treatment resistance in pancreatic Cancer. Cancers (Basel) 9 (2017)
A.D. Rhim, E.T. Mirek, N.M. Aiello, A. Maitra, J.M. Bailey, F. Mcallister, G.L. Beatty, A.K. Rustgi, R.H. Vonderheide, S.D. Leach, B.Z. Stanger, EMT and dissemination precede pancreatic tumor formation. Cell 148, 349–361 (2012)
D.M. Gonzalez, D. Medici, Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 7, re8 (2014)
J.M. Lee, S. Dedhar, R. Kalluri, E.W. Thompson, The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J. Cell Biol. 172, 973–981 (2006)
M. Nakamoto, M. Hisaoka, Clinicopathological implications of wingless/int1 (WNT) signaling pathway in pancreatic ductal adenocarcinoma. J. UOEH 38, 1–8 (2016)
W.H. Lien, E. Fuchs, Wnt some lose some: Transcriptional governance of stem cells by Wnt/β-catenin signaling. Genes Dev. 28, 1517–1532 (2014)
V.S. Li, S.S. Ng, P.J. Boersema, T.Y. Low, W.R. Karthaus, J.P. Gerlach, S. Mohammed, A.J.R. Heck, M.M. Maurice, T. Mahmoudi, H. Cleverset, Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell 149, 1245–1256 (2012)
L.C. Platanias, Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 (2005)
R.C. Fleith, H.V. Mears, X.Y. Leong, T.J. Sanford, E. Emmott, S.C. Graham, D. S. Mansur, T.R. Sweeney, IFIT3 and IFIT2/3 promote IFIT1-mediated translation inhibition by enhancing binding to non-self RNA. Nucleic Acids Res. 46, 5269–5285 (2018)
V.K. Pidugu, M.M. Wu, A.H. Yen, H.B. Pidugu, K.W. Chang, C.J. Liu, T.-C. Lee, IFIT1 and IFIT3 promote oral squamous cell carcinoma metastasis and contribute to the anti-tumor effect of gefitinib via enhancing p-EGFR recycling. Oncogene 38, 3232–3247 (2019)
G. Liu, J. Sun, Z.F. Yang, C. Zhou, P.Y. Zhou, R.Y. Guan, B.-Y. Sun, Z.-T. Wang, J. Zhou, J. Fan, S.-J. Qiu, Y. Yi, Cancer-associated fibroblast-derived CXCL11 modulates hepatocellular carcinoma cell migration and tumor metastasis through the circUBAP2/miR-4756/IFIT1/3 axis. Cell Death Dis. 12, 260 (2021)
R.L. Siegel, K.D. Miller, A. Jemal, Cancer statistics, 2019. CA Cancer J. Clin. 69, 7–34 (2019)
S.H. Lin, G.S. Raju, C. Huff, Y. Ye, J. Gu, J.S. Chen, M.A.T. Hildebrandt, H. Liang, D.G. Menter, J. Morris, E. Hawk, J.R. Stroehlein, A. Futreal, S. Kopetz, L. Mishra, X. Wu., The somatic mutation landscape of premalignant colorectal adenoma. Gut 67, 1299–1305 (2018)
N. Marschner, S. Zacharias, F. Lordick, S. Hegewisch-Becker, U. Martens, A. Welt, V. Hagen, W. Gleiber, S. Bohnet, L. Kruggel, S. Dille, A. Nusch, S. Dörfel, T. Decker, M. Jänicke, Association of Disease Progression with Health-Related Quality of life among adults with breast, lung, pancreatic, and colorectal cancer. JAMA Netw. Open 3, e200643 (2020)
D. Creytens, NKX2.2 immunohistochemistry in the distinction of Ewing sarcoma from cytomorphologic mimics: Diagnostic utility and pitfalls-comment on Russell-Goldman et al. Cancer Cytopathol. 127, 202 (2019)
B.Q. Li, Z.Y. Liang, S. Seery, Q.F. Liu, L. You, T.P. Zhang, J.C. Guo, Y.P. Zhao, WT1 associated protein promotes metastasis and chemo-resistance to gemcitabine by stabilizing Fak mRNA in pancreatic cancer. Cancer Lett. 451, 48–57 (2019)
K. Willert, R. Nusse, Beta-catenin: A key mediator of Wnt signaling. Curr. Opin. Genet. Dev. 8, 95–102 (1998)
S. Yang, Y. Liu, M.Y. Li, C.S.H. Ng, S.L. Yang, S. Wang, C. Zou, Y. Dong, J. Du, X. Long, L.-Z. Liu, I.Y.P. Wan, T. Mok, M.J. Underwood, G.G. Chen, FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol. Cancer 16, 124 (2017)
C.C. Liu, D.L. Cai, F. Sun, Z.H. Wu, B. Yue, S.L. Zhao, X.-S. Wu, M. Zhang, X.-W. Zhu, Z.-H. Peng, D.-W. Yan, FERMT1 mediates epithelial-mesenchymal transition to promote colon cancer metastasis via modulation of β-catenin transcriptional activity. Oncogene 36, 1779–1792 (2017)
V. Fensterl, G.C. Sen, Interferon-induced Ifit proteins: Their role in viral pathogenesis. J. Virol. 89, 2462–2468 (2015)
R.K. Allan, T. Ratajczak, Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones 16, 353–367 (2011)
V. Fensterl, G.C. Sen, The ISG56/IFIT1 gene family. J. Interf. Cytokine Res. 31, 71–78 (2011)
H.H. Danish, S. Goyal, N.K. Taunk, H. Wu, M.S. Moran, B.G. Haffty, Interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) as a prognostic marker for local control in T1-2 N0 breast cancer treated with breast-conserving surgery and radiation therapy (BCS + RT). Breast J. 19, 231–239 (2013)
Y. Zhao, A. Altendorf-Hofmann, I. Pozios, P. Camaj, T. Däberitz, X. Wang, H. Niess, H. Seeliger, F. Popp, C. Betzlerr, U. Settmacher, K.-W. Jauch, C. Bruns, T. Knösel, Elevated interferon-induced protein with tetratricopeptide repeats 3 (IFIT3) is a poor prognostic marker in pancreatic ductal adenocarcinoma. J. Cancer Res. Clin. Oncol. 143, 1061–1068 (2017)
Y. Zhang, Y. Kong, S. Liu, L. Zeng, L. Wan, Z. Zhang, Curcumin induces apoptosis in human leukemic cell lines through an IFIT2-dependent pathway. Cancer Biol. Ther. 18, 43–50 (2017)
Y. Yang, Y. Zhou, J. Hou, C. Bai, Z. Li, J. Fan, I.O. L. Ng, W. Zhou, H. Sun, Q. Dong, J.M.F. Lee, C.-M. Lo, K. Man, Y. Yang, N. Li, G. Ding, Y. Yu, X. Cao, Hepatic IFIT3 predicts interferon-α therapeutic response in patients of hepatocellular carcinoma. Hepatology 66, 152–166 (2017)
M.A. Nieto, R.Y. Huang, R.A. Jackson, J.P. Thiery, EMT: 2016. Cell 166, 21–45 (2016)
H. Hugo, M.L. Ackland, T. Blick, M.G. Lawrence, J.A. Clements, E.D. Williams, E.W. Thompsonet, Epithelial--mesenchymal and mesenchymal--epithelial transitions in carcinoma progression. J. Cell. Physiol. 213, 374–383 (2007)
S. Valastyan, R.A. Weinberg, Tumor metastasis: Molecular insights and evolving paradigms. Cell 147, 275–292 (2011)
A. Puisieux, T. Brabletz, J. Caramel, Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 16, 488–494 (2014)
Acknowledgements
This work was supported by the Natural Science Foundation of China (No. 81773215) and the Chinese Academy of Medical Sciences (No. 2019XK320002).
Author information
Authors and Affiliations
Contributions
LTH and ZBB contributed to the research design, preparation of the manuscript and collection of the data. QC, WYY, LZR, YXY and ZXT designed the research and revised the manuscript. WWB supervised the research. All the authors contributed to the article and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All the authors have no conflicts of interest to declare.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tian-Hao Li and Bang-Bo Zhao are co-first authors.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1007/s13402-023-00897-4"
Supplementary Information
Supplementary Figure 1
mRNA expression of IFIT1 in pancreatic cancer cell lines. Transient transduction of IFIT1 knockdown and overexpression constructs was performed, and the mRNA expression of β-catenin was examined. (A) mRNA expression of IFIT1 in 6 PC cell lines. (B) The efficiency of IFIT1 KD in Aspc-1 and Bxpc-3 cells was confirmed by qPCR. (C) The efficiency of IFIT1 overexpression in Aspc-1 and Panc-1 cells was confirmed by qPCR. (D, E) The mRNA expression of β-catenin in transiently transduced cells. (JPG 1107 kb)
Supplementary Table 1
Correlation between IFIT1 expression and clinicopathological characteristics of pancreatic cancer patients in TCGA datasets. (JPG 1107 kb) (DOCX 20 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, TH., Zhao, BB., Qin, C. et al. RETRACTED ARTICLE: IFIT1 modulates the proliferation, migration and invasion of pancreatic cancer cells via Wnt/β-catenin signaling. Cell Oncol. 44, 1425–1437 (2021). https://doi.org/10.1007/s13402-021-00651-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-021-00651-8