Abstract
Background
Previously, we found that the Graffi murine leukemia virus (MuLV) is able to induce a wide spectrum of hematologic malignancies in vivo. Using high-density oligonucleotide microarrays, we established the gene expression profiles of several of these malignancies, thereby specifically focusing on genes deregulated in the lymphoid sub-types. We observed over-expression of a variety of genes, including Arntl2, Bfsp2, Gfra2, Gpm6a, Gpm6b, Nln, Fbln1, Bmp7, Etv5 and Celsr1 and, in addition, provided evidence that Fmn2 and Parm-1 may act as novel oncogenes. In the present study, we assessed the expression patterns of eight selected human homologs of these genes in primary human B-cell malignancies, and explored the putative oncogenic potential of GPM6A and GPM6B.
Methods
The gene expression levels of the selected human homologs were tested in human B-cell malignancies by semi-quantitative RT-PCR. The protein expression profiles of human GPM6A and GPM6B were analyzed by Western blotting. The localization and the effect of GPM6A and GPM6B on the cytoskeleton were determined using confocal and indirect immunofluorescence microscopy. To confirm the oncogenic potential of GPM6A and GPM6B, classical colony formation assays in soft agar and focus forming assays were used. The effects of these proteins on the cell cycle were assessed by flow cytometry analysis.
Results
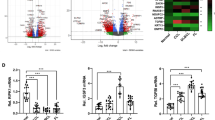
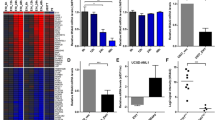
Using semi-quantitative RT-PCR, we found that most of the primary B-cell malignancies assessed showed altered expression patterns of the genes tested, including GPM6A and GPM6B. Using confocal microscopy, we found that the GPM6A protein (isoform 3) exhibits a punctate cytoplasmic localization and that the GPM6B protein (isoform 4) exhibits a peri-nuclear and punctate cytoplasmic localization. Interestingly, we found that exogenous over-expression of both proteins in NIH/3T3 cells alters the actin and microtubule networks and induces the formation of long filopodia-like protrusions. Additionally, we found that these over-expressing NIH/3T3 cells exhibit anchorage-independent growth and enhanced proliferation rates. Cellular transformation (i.e., loss of contact inhibition) was, however, only observed after exogenous over-expression of GPM6B.
Conclusions
Our results indicate that several human homologs of the genes found to be deregulated in Graffi MuLV experimental mouse models may serve as candidate biomarkers for human B-cell malignancies. In addition, we found that GPM6A and GPM6B may act as novel oncogenes in the development of these malignancies.





Similar content being viewed by others
References
A. Graffi, Chloroleukemia of mice. Ann. N. Y. Acad. Sci. 68, 540–558 (1957)
V. Voisin, C. Barat, T. Hoang, E. Rassart, Novel insights into the pathogenesis of the Graffi murine leukemia retrovirus. J. Virol. 80, 4026–4037 (2006)
C. Denicourt, E. Edouard, E. Rassart, Oncogene activation in myeloid leukemias by Graffi murine leukemia virus proviral integration. J. Virol. 73, 4439–4442 (1999)
C. Denicourt, C.A. Kozak, E. Rassart, Gris1, a new common integration site in Graffi murine leukemia virus-induced leukemias: overexpression of a truncated cyclin D2 due to alternative splicing. J. Virol. 77, 37–44 (2003)
C. Denicourt, P. Legault, F.A. McNabb, E. Rassart, Human and mouse cyclin D2 splice variants: transforming activity and subcellular localization. Oncogene 27, 1253–1262 (2008)
C. Charfi, V. Voisin, L.C. Levros Jr., E. Edouard, E. Rassart, Gene profiling of Graffi murine leukemia virus-induced lymphoid leukemias: identification of leukemia markers and Fmn2 as a potential oncogene. Blood 117, 1899–1910 (2011)
C. Charfi, L.C. Levros Jr., E. Edouard, E. Rassart, Characterization and identification of PARM-1 as a new potential oncogene. Mol. Cancer 12, 84 (2013)
X. Castells, J.J. Acebes, S. Boluda, A. Moreno-Torres, J. Pujol, M. Julia-Sape, A.P. Candiota, J. Arino, A. Barcelo, C. Arus, Development of a predictor for human brain tumors based on gene expression values obtained from two types of microarray technologies. OMICS 14, 157–164 (2010)
P. Urban, M. Bilecova-Rabajdova, Z. Stefekova, A. Ostro, M. Marekova, Overview of potential oncomarkers for detection of early stages of ovarian cancer. Klin. Onkol. 24, 106–111 (2011)
S. Landais, S. Landry, P. Legault, E. Rassart, Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 67, 5699–5707 (2007)
M. Ru, C. Shustik, E. Rassart, Graffi murine leukemia virus: molecular cloning and characterization of the myeloid leukemia-inducing agent. J. Virol. 67, 4722–4731 (1993)
E. Edouard, C. Charfi, V. Voisin, C. Denicourt, C. Barat, E. Rassart, The Graffi murine leukemia virus: a multipotent retrovirus ideal for the study and characterization of lymphoid and non lymphoid leukemias. Transw. Res. Netw. 8, 23–47 (2012)
V. Voisin, P. Legault, D.P. Ospina, Y. Ben-David, E. Rassart, Gene profiling of the erythro- and megakaryoblastic leukaemias induced by the Graffi murine retrovirus. BMC Med. Genomics 3, 2 (2010)
F. Sievers, A. Wilm, D. Dineen, T.J. Gibson, K. Karplus, W. Li, R. Lopez, H. McWilliam, M. Remmert, J. Soding, J.D. Thompson, D.G. Higgins, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011)
K.C. Chou, H.B. Shen, Signal-CF: a subsite-coupled and window-fusing approach for predicting signal peptides. Biochem. Biophys. Res. Commun. 357, 633–640 (2007)
N.H. Colburn, W.F. Bruegge, J.R. Bates, R.H. Gray, J.D. Rossen, W.H. Kelsey, T. Shimada, Correlation of anchorage-independent growth with tumorigenicity of chemically transformed mouse epidermal cells. Cancer Res. 38, 624–634 (1978)
V.H. Freedman, S.I. Shin, Cellular tumorigenicity in nude mice: correlation with cell growth in semi-solid medium. Cell 3, 355–359 (1974)
S.I. Shin, V.H. Freedman, R. Risser, R. Pollack, Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc. Natl. Acad. Sci. U. S. A. 72, 4435–4439 (1975)
C. Charfi, E. Edouard, E. Rassart, Lymphoid leukemias induced by the murine retrovirus Graffi. http://www.biomed.uqam.ca/rassart/microarray2.html. Accessed 14 November 2013
M. Ikeda, W. Yu, M. Hirai, T. Ebisawa, S. Honma, K. Yoshimura, K.I. Honma, M. Nomura, cDNA cloning of a novel bHLH-PAS transcription factor superfamily gene, BMAL2: its mRNA expression, subcellular distribution, and chromosomal localization. Biochem. Biophys. Res. Commun. 275, 493–502 (2000)
S. Jones, An overview of the basic helix-loop-helix proteins. Genome Biol. 5, 226 (2004)
Y. Yasuniwa, H. Izumi, K.Y. Wang, S. Shimajiri, Y. Sasaguri, K. Kawai, H. Kasai, T. Shimada, K. Miyake, E. Kashiwagi, G. Hirano, A. Kidani, M. Akiyama, B. Han, Y. Wu, I. Ieiri, S. Higuchi, K. Kohno, Circadian disruption accelerates tumor growth and angio/stromagenesis through a Wnt signaling pathway. PLoS ONE 5, e15330 (2010)
G. Mazzoccoli, V. Pazienza, A. Panza, M.R. Valvano, G. Benegiamo, M. Vinciguerra, A. Andriulli, A. Piepoli, ARNTL2 and SERPINE1: potential biomarkers for tumor aggressiveness in colorectal cancer. J. Cancer Res. Clin. Oncol. 138, 501–511 (2012)
S.D. Georgatos, F. Gounari, S. Remington, The beaded intermediate filaments and their potential functions in eye lens. BioEssays 16, 413–418 (1994)
M.D. Perng, Q. Zhang, R.A. Quinlan, Insights into the beaded filament of the eye lens. Exp. Cell Res. 313, 2180–2188 (2007)
M.A. Aldahmesh, A.O. Khan, J. Mohamed, F.S. Alkuraya, Novel recessive BFSP2 and PITX3 mutations: insights into mutational mechanisms from consanguineous populations. Genet. Med. 13, 978–981 (2011)
G. Wu, L. Zhou, L. Khidr, X.E. Guo, W. Kim, Y.M. Lee, T. Krasieva, P.L. Chen, A novel role of the chromokinesin Kif4A in DNA damage response. Cell Cycle 7, 2013–2020 (2008)
Y. Kurasawa, W.C. Earnshaw, Y. Mochizuki, N. Dohmae, K. Todokoro, Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 23, 3237–3248 (2004)
M. Mazumdar, J.H. Lee, K. Sengupta, T. Ried, S. Rane, T. Misteli, Tumor formation via loss of a molecular motor protein. Curr. Biol. 16, 1559–1564 (2006)
Y.S. Kim, J.D. Hwan, S. Bae, D.H. Bae, W.A. Shick, Identification of differentially expressed genes using an annealing control primer system in stage III serous ovarian carcinoma. BMC Cancer 10, 576 (2010)
L.L. Yates, C. Schnatwinkel, J.N. Murdoch, D. Bogani, C.J. Formstone, S. Townsend, A. Greenfield, L.A. Niswander, C.H. Dean, The PCP genes Celsr1 and Vangl2 are required for normal lung branching morphogenesis. Hum. Mol. Genet. 19, 2251–2267 (2010)
M. Katoh, Comparative integromics on non-canonical WNT or planar cell polarity signaling molecules: transcriptional mechanism of PTK7 in colorectal cancer and that of SEMA6A in undifferentiated ES cells. Int. J. Mol. Med. 20, 405–409 (2007)
C. Korz, A. Pscherer, A. Benner, D. Mertens, C. Schaffner, E. Leupolt, H. Dohner, S. Stilgenbauer, P. Lichter, Evidence for distinct pathomechanisms in B-cell chronic lymphocytic leukemia and mantle cell lymphoma by quantitative expression analysis of cell cycle and apoptosis-associated genes. Blood 99, 4554–4561 (2002)
A.W. Fjorback, H.K. Muller, O. Wiborg, Membrane glycoprotein M6B interacts with the human serotonin transporter. J. Mol. Neurosci. 37, 191–200 (2009)
S. Olinsky, B.T. Loop, A. DeKosky, B. Ripepi, W. Weng, J. Cummins, S.L. Wenger, Y. Yan, C. Lagenaur, V. Narayanan, Chromosomal mapping of the human M6 genes. Genomics 33, 532–536 (1996)
N.L. Nadon, S. Miller, K. Draeger, M. Salvaggio, Myelin proteolipid DM20: evidence for function independent of myelination. Int. J. Dev. Neurosci. 15, 285–293 (1997)
X. Castells, J.M. Garcia-Gomez, A. Navarro, J.J. Acebes, O. Godino, S. Boluda, A. Barcelo, M. Robles, J. Arino, C. Arus, Automated brain tumor biopsy prediction using single-labeling cDNA microarrays-based gene expression profiling. Diagn. Mol. Pathol. 18, 206–218 (2009)
Y. Yan, C. Lagenaur, V. Narayanan, Molecular cloning of M6: identification of a PLP/DM20 gene family. Neuron 11, 423–431 (1993)
H. Werner, L. Dimou, M. Klugmann, S. Pfeiffer, K.A. Nave, Multiple splice isoforms of proteolipid M6B in neurons and oligodendrocytes. Mol. Cell. Neurosci. 18, 593–605 (2001)
J.L. Popot, D. Pham Dinh, A. Dautigny, Major myelin proteolipid: the 4-alpha-helix topology. J. Membr. Biol. 120, 233–246 (1991)
T. Weimbs, W. Stoffel, Proteolipid protein (PLP) of CNS myelin: positions of free, disulfide-bonded, and fatty acid thioester-linked cysteine residues and implications for the membrane topology of PLP. Biochemistry 31, 12289–12296 (1992)
A. Gow, A. Gragerov, A. Gard, D.R. Colman, R.A. Lazzarini, Conservation of topology, but not conformation, of the proteolipid proteins of the myelin sheath. J. Neurosci. 17, 181–189 (1997)
A.M. Schneider, I.R. Griffiths, C. Readhead, K.A. Nave, Dominant-negative action of the jimpy mutation in mice complemented with an autosomal transgene for myelin proteolipid protein. Proc. Natl. Acad. Sci. U. S. A. 92, 4447–4451 (1995)
E. Swanton, A. Holland, S. High, P. Woodman, Disease-associated mutations cause premature oligomerization of myelin proteolipid protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 102, 4342–4347 (2005)
S. Yang, W. Wang, C. Lei, Q. Liu, F. Xu, X. Xing, H. Chen, J. Liu, S. Wu, M. Wang, Localization and characterization of rat transmembrane protein 225 specifically expressed in testis. DNA Cell Biol. 30, 9–16 (2011)
M. Namiki, M. Kitamura, E. Buczko, M.L. Dufau, Rat testis P-450(17)alpha cDNA: the deduced amino acid sequence, expression and secondary structural configuration. Biochem. Biophys. Res. Commun. 157, 705–712 (1988)
J. Haugstetter, T. Blicher, L. Ellgaard, Identification and characterization of a novel thioredoxin-related transmembrane protein of the endoplasmic reticulum. J. Biol. Chem. 280, 8371–8380 (2005)
M. Schweneker, A.S. Bachmann, K. Moelling, JM4 is a four-transmembrane protein binding to the CCR5 receptor. FEBS Lett. 579, 1751–1758 (2005)
M. Wang, G. Luo, F. Li, S. Wu, J. Jiang, C. Huang, Isolation and characterization of a novel four-transmembrane protein PMP22CD specifically expressed in the testis. Int. J. Mol. Sci. 7, 425–437 (2006)
H. Ohno, J. Stewart, M.C. Fournier, H. Bosshart, I. Rhee, S. Miyatake, T. Saito, A. Gallusser, T. Kirchhausen, J.S. Bonifacino, Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science 269, 1872–1875 (1995)
J.S. Bonifacino, E.C. Dell’Angelica, Molecular bases for the recognition of tyrosine-based sorting signals. J. Cell Biol. 145, 923–926 (1999)
B.D. Trapp, A. Nishiyama, D. Cheng, W. Macklin, Molecular bases for the recognition of tyrosine-based sorting signals. J. Cell Biol. 137, 459–468 (1997)
M.P. Sinoway, K. Kitagawa, S. Timsit, G.A. Hashim, D.R. Colman, Proteolipid protein interactions in transfectants: implications for myelin assembly. J. Neurosci. Res. 37, 551–562 (1994)
J. Alfonso, M.E. Fernandez, B. Cooper, G. Flugge, A.C. Frasch, The stress-regulated protein M6a is a key modulator for neurite outgrowth and filopodium/spine formation. Proc. Natl. Acad. Sci. U. S. A. 102, 17196–17201 (2005)
B. Fuchsova, M.E. Fernandez, J. Alfonso, A.C. Frasch, Cysteine residues in the large extracellular loop (EC2) are essential for the function of the stress-regulated glycoprotein M6a. J. Biol. Chem. 284, 32075–32088 (2009)
M.A. Brocco, M.E. Fernandez, A.C. Frasch, Filopodial protrusions induced by glycoprotein M6a exhibit high motility and aids synapse formation. Eur. J. Neurosci. 31, 195–202 (2010)
K.Y. Huang, G.D. Chen, C.H. Cheng, K.Y. Liao, C.C. Hung, G.D. Chang, P.P. Hwang, S.Y. Lin, M.C. Tsai, K.H. Khoo, M.T. Lee, C.J. Huang, Phosphorylation of the zebrafish M6Ab at serine 263 contributes to filopodium formation in PC12 cells and neurite outgrowth in zebrafish embryos. PLoS ONE 6, e26461 (2011)
M.E. Fernandez, J. Alfonso, M.A. Brocco, A.C. Frasch, Conserved cellular function and stress-mediated regulation among members of the proteolipid protein family. J. Neurosci. Res. 88, 1298–1308 (2010)
F. Xue, D.M. Janzen, D.A. Knecht, Contribution of filopodia to cell migration: a mechanical link between protrusion and contraction. Int. J. Cell Biol. 2010, 507821 (2010)
A. Arjonen, R. Kaukonen, J. Ivaska, Filopodia and adhesion in cancer cell motility. Cell Adhes. Migr. 5, 421–430 (2011)
H. Li, S. Hou, X. Wu, S. Nandagopal, F. Lin, S. Kung, A.J. Marshall, The tandem PH domain-containing protein 2 (TAPP2) regulates chemokine-induced cytoskeletal reorganization and malignant B cell migration. PLoS ONE 8, e57809 (2013)
C. Scielzo, M.T. Bertilaccio, G. Simonetti, A. Dagklis, E. ten Hacken, C. Fazi, M. Muzio, V. Caiolfa, D. Kitamura, U. Restuccia, A. Bachi, M. Rocchi, M. Ponzoni, P. Ghia, F. Caligaris-Cappio, HS1 has a central role in the trafficking and homing of leukemic B cells. Blood 116, 3537–3546 (2010)
Acknowledgments
We thank Dr. Daniel Sinnet for providing the pediatric tumor samples, Bertrand Fournier for his help with the statistical analysis and Denis Flipo for his help with the confocal microscopy analyses. This work was supported by the Canadian Institutes of Health Research, grant MOP-37994 (ER). CC is a recipient of a studentship from the Tunisia Government and Fondation UQAM.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource Fig. 1
Alignment of deduced amino acid sequences for GPM6A isoforms together and for GPM6B isoforms together. Alignment a of the 5 Homo sapiens isoforms of GPM6A (NP_005268 (isoform 1), NP_963885 (isoform 1), NP_001248376 (isoform 2), NP_963886 (isoform 3) and NP_001248377 (isoform 4)) and b of the 4 homosapiens isoforms of GPM6B protein (NP_001001995 (isoform 1), NP_005269 (isoform 3), NP_001001996 (isoform 2) and NP_001001994 (isoform 4)) using the Clustal Omega software. Stars indicate a perfect match between all aligned isoforms and dashed lines indicate gaps. (PDF 30 kb)
Online Resource Fig. 2
Alignment of GPM6A, GPM6B and DM20 proteins. The amino acid residues of GPM6A isoform 3, GPM6B isoform 4 and DM20 proteins were aligned in pairs to determine their identity percentage. a Alignment of GPM6A isoform 3 and GPM6B isoform 4. b Alignment of DM20 and GPM6A isoform 3. c Alignment of DM20 and GPM6B isoform 4. Boxes: N- and C- cytoplasmic tails, dotted lines: transmembrane domains, lines: extracellular loops, double lines: intracellular loop. (PDF 48 kb)
Rights and permissions
About this article
Cite this article
Charfi, C., Edouard, E. & Rassart, E. Identification of GPM6A and GPM6B as potential new human lymphoid leukemia-associated oncogenes. Cell Oncol. 37, 179–191 (2014). https://doi.org/10.1007/s13402-014-0171-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13402-014-0171-y