Abstract
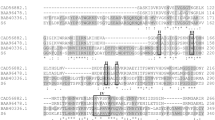
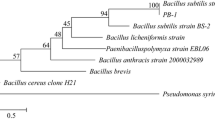
Pectinases have broad applications in food industries attributed to their role as a biocatalyst in degrading pectic compounds. The present work reports the purification and characterization of a pectinase from a mutant strain of Bacillus subtilis. A three-step enzyme purification involving ammonium sulfate precipitation, followed by ion-exchange chromatography (IEC) and size-exclusion chromatography (SEC), was evaluated to recover the enzyme effectively. The pectinase was purified up to 7.98-fold with 65.63% total recovery, resulting in maximum specific activity of 362.04 U/mg after three purification steps. The eluted fractions of IEC and SEC showed pectinase activities of 9520 U and 8288 U, respectively. The molecular weight of this extracellular pectinase was found to be 30 kDa after sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis. The optimal conditions of temperature and pH for maximum pectinase activity were 37 °C and 4–5, respectively. The enzyme also exhibited high stability when incubated at optimal conditions for 60 min. The addition of calcium and magnesium ions elicited pectinase activity at 4 mM concentrations, while copper and zinc ions inhibited the pectinase activity. The Vmax and Km values were 362 U and 0.62 mg/ml, respectively, for the pectin substrate obtained from citrus peel. Further analysis on the efficiency of clarification in sweet lime, pineapple, and lime juice showed 79.25, 72.76, and 61.24% reductions in turbidity within 1-h incubation. Therefore, characteristics of pectinase suggest that it can be potentially used for industrial applications as a biocatalyst.









Similar content being viewed by others
References
Kc S, Upadhyaya J, Joshi DR, Lekhak B, Kumar Chaudhary D, Raj Pant B, Raj Bajgai T, Dhital R, Khanal S, Koirala N (2020) Production, characterization, and industrial application of pectinase enzyme isolated from fungal strains. Fermentation 6(2):59–69. https://doi.org/10.3390/fermentation6020059
Cornuault V, Posé S, Knox JP (2018) Disentangling pectic homogalacturonan and rhamnogalacturonan-I polysaccharides: evidence for sub-populations in fruit parenchyma systems. Food Chem 246:275–285. https://doi.org/10.1016/j.foodchem.2017.11.025
John J, Kaimal KS, Smith ML, Rahman PK, Chellam PV (2020) Advances in upstream and downstream strategies of pectinase bioprocessing: a review. Int J Biol Macromol 162:1086–1099. https://doi.org/10.1016/j.ijbiomac.2020.06.224
Rebello S, Anju M, Aneesh EM, Sindhu R, Binod P, Pandey A (2017) Recent advancements in the production and application of microbial pectinases: an overview. Reviews in environmental science and bio/technology 16(3):381–394. https://doi.org/10.1007/s11157-017-9437-y
Khatri BP, Bhattarai T, Shrestha S, Maharjan J (2015) Alkaline thermostable pectinase enzyme from Aspergillus niger strain MCAS2 isolated from Manaslu Conservation Area, Gorkha. Nepal SpringerPlus 4(1):1–8. https://doi.org/10.1186/s40064-015-1286-y
El Enshasy HA, Elsayed EA, Suhaimi N, Abd Malek R, Esawy M (2018) Bioprocess optimization for pectinase production using Aspergillus niger in a submerged cultivation system. BMC Biotechnol 18(1):1–13. https://doi.org/10.1186/s12896-018-0481-7
Lima TCSd, Grisi BM, Bonato MCM (1999) Bacteria isolated from a sugarcane agroecosystem: their potential production of polyhydroxyalcanoates and resistance to antibiotics. Rev Microbiol 30(3):214–224. https://doi.org/10.1590/S0001-37141999000300006
Ying-xian Y (2005) The status quo and strategy of hemp bast degumming. Shandong Textile Science & Technology 1:51–54. https://doi.org/10.1590/CJFDTotal-SDKJ200501018
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. https://doi.org/10.1006/abio.1976.9999
Bhunia B, Basak B, Mandal T, Bhattacharya P, Dey A (2013) Effect of pH and temperature on stability and kinetics of novel extracellular serine alkaline protease (70 kDa). Int J Biol Macromol 54:1–8. https://doi.org/10.1016/j.ijbiomac.2012.11.024
Uday USP, Majumdar R, Tiwari ON, Mishra U, Mondal A, Bandyopadhyay TK, Bhunia B (2017) Isolation, screening and characterization of a novel extracellular xylanase from Aspergillus niger (KP874102. 1) and its application in orange peel hydrolysis. Int J Biol Macromol 105:401–409. https://doi.org/10.1016/j.ijbiomac.2017.07.066
Salazar N, Ruas-Madiedo P, Prieto A, Calle LP, de los Reyes-Gavilán CG, (2012) Characterization of exopolysaccharides produced by Bifidobacterium longum NB667 and its cholate-resistant derivative strain IPLA B667dCo. J Agric Food Chem 60(4):1028–1035. https://doi.org/10.1021/jf204034n
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259):680–685. https://doi.org/10.1038/227680a0
Mukesh K, Saranya GM, Suresh K, Andal P, Rajakumar R, Kalaichelvan P (2012) Production and optimization of pectinase from Bacillus sp. MFW7 using cassava waste. Asian Journal of Plant Science and Research 2(3):369–375. https://doi.org/10.1341/aj204034
Bhunia B, Dutta D, Chaudhuri S (2011) Extracellular alkaline protease from Bacillus licheniformis NCIM-2042: Improving enzyme activity assay and characterization. Eng Life Sci 11:207–215. https://doi.org/10.1002/elsc.201000020
Bajaj BK, Manhas K (2012) Production and characterization of xylanase from Bacillus licheniformis P11 (C) with potential for fruit juice and bakery industry. Biocatal Agric Biotechnol 1(4):330–337. https://doi.org/10.1016/j.bcab.2012.07.003
Su Y, Liu C, Fang H, Zhang D (2020) Bacillus subtilis: a universal cell factory for industry, agriculture, biomaterials and medicine. Microb Cell Fact 19(1):1–12. https://doi.org/10.1186/s12934-020-01436-8
Yu P, Xu C (2018) Production optimization, purification and characterization of a heat-tolerant acidic pectinase from Bacillus sp. ZJ1407. Int J Biol Macromol 108:972–980. https://doi.org/10.1016/j.ijbiomac.2017.11.012
Oumer OJ, Abate D (2017) Characterization of pectinase from Bacillus subtilis strain Btk 27 and its potential application in removal of mucilage from coffee beans. Enzyme Res 2017:1–7. https://doi.org/10.1155/2017/7686904
Ahlawat S, Mandhan R, Dhiman SS, Kumar R, Sharma J (2008) Potential application of alkaline pectinase from Bacillus subtilis SS in pulp and paper industry. Appl Biochem Biotechnol 149(3):287–293. https://doi.org/10.1007/s12010-007-8096-9
Rahman MS, Kim YK, Khan MM, Lee SH, Choi YH, Cho SS, Park C, Yoo JC (2020) Purification and identification of novel alkaline pectinase PNs31 from Bacillus subtilis CBS31 and its immobilization for bioindustrial applications. Korean J Chem Eng 37(11):1942–1950. https://doi.org/10.1007/s11814-020-0648-5
Ahmed I, Zia MA, Hussain MA, Akram Z, Naveed MT, Nowrouzi A (2016) Bioprocessing of citrus waste peel for induced pectinase production by Aspergillus niger; its purification and characterization. Journal of radiation research and applied sciences 9(2):148–154. https://doi.org/10.1016/j.jrras.2015.11.003
Prajapati J, Dudhagara P, Patel K (2021) Production of thermal and acid-stable pectinase from Bacillus subtilis strain BK-3: optimization, characterization, and application for fruit juice clarification. Biocatal Agric Biotechnol 35:102063. https://doi.org/10.1016/j.bcab.2021.102063
Tiwari ON, Mondal A, Bhunia B, kanti Bandyopadhyay T, Jaladi P, Oinam G, Indrama T (2019) Purification, characterization and biotechnological potential of new exopolysaccharide polymers produced by cyanobacterium Anabaena sp. CCC 745. Polymer 178:121695. https://doi.org/10.1016/j.polymer.2019.121695
Jayani RS, Saxena S, Gupta R (2005) Microbial pectinolytic enzymes: a review. Process Biochem 40(9):2931–2944. https://doi.org/10.1016/j.procbio.2005.03.026
Ma X, Chen W, Yan T, Wang D, Hou F, Miao S, Liu D (2020) Comparison of citrus pectin and apple pectin in conjugation with soy protein isolate (SPI) under controlled dry-heating conditions. Food Chem 309:125501. https://doi.org/10.1016/j.foodchem.2019.125501
Saad N, Briand M, Gardarin C, Briand Y, Michaud P (2007) Production, purification and characterization of an endopolygalacturonase from Mucor rouxii NRRL 1894. Enzyme Microb Technol 41(6–7):800–805. https://doi.org/10.1016/j.enzmictec.2007.07.012
Dixon M, Webb EC (1979) Enzymes. 3rd edn. Academic Press, New York. https://doi.org/10.1016/978-0-12-122703-6
Tanford C (1968) Protein denaturation Adv Prot Chem 23:121–282. https://doi.org/10.1016/s0065-3233(08)60401-5
Sadana A, Henley JP (1988) Influence of pH on enzyme stabilization:an analysis using a series-type mechanism. J Biotechnol 7:95–112. https://doi.org/10.1016/0168-1656(88)90057-0
Sode K, Ito K, Witarto AB, Watanabe K, Yoshida H, Postma P (1996) Increased production of recombinant pyrroloquinoline quinone (PQQ) glucose dehydrogenase by metabolically engineered Escherichia coli strain capable of PQQ biosynthesis. J Biotechnol 49(1–3):239–243. https://doi.org/10.1016/0168-1656(96)01540-4
Butt MS, Tahir-Nadeem M, Ahmad Z, Sultan MT (2008) Xylanases and their applications in baking industry. Food Technology and Biotechnology 46 (1):22–31. https://doi.org/10.17113/ftb.56.01.18.5477
Acknowledgements
Ram Balak Mahto and Mukesh Yadav express gratitude to HoD, Department of Biotechnology, Maharishi Markandeshwar (Deemed to be University), Mullana 133207, Ambala, India, for their encouragement and support. Dr. Biswanath Bhunia and Muthusivaramapandian Muthuraj express gratitude to the Director, National Institute of Technology Agartala for the constant inspiration and funding.
Funding
Dr. Biswanath Bhunia would like to express their gratitude to the Department of Science and Technology (DST), Government of India, for providing financial assistance to Fast Track Young Scientist (SERC/LS-167/2012).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mahto, R.B., Yadav, M., Muthuraj, M. et al. Biochemical properties and application of a novel pectinase from a mutant strain of Bacillus subtilis. Biomass Conv. Bioref. 13, 10463–10474 (2023). https://doi.org/10.1007/s13399-021-02225-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13399-021-02225-y