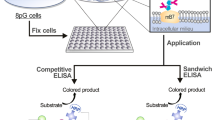
Abstract
We describe epitope mapping data using multiple covalent labeling footprinting-mass spectrometry (MS) techniques coupled with negative stain transmission electron microscopy (TEM) data to analyze the antibody–antigen interactions in a sandwich enzyme-linked immunosorbant assay (ELISA). Our hydroxyl radical footprinting-MS data using fast photochemical oxidation of proteins (FPOP) indicates suppression of labeling across the antigen upon binding either of the monoclonal antibodies (mAbs) utilized in the ELISA. Combining these data with Western blot analysis enabled the identification of the putative epitopes that appeared to span regions containing N-linked glycans. An additional structural mapping technique, carboxyl group footprinting-mass spectrometry using glycine ethyl ester (GEE) labeling, was used to confirm the epitopes. Deglycosylation of the antigen resulted in loss of potency in the ELISA, supporting the FPOP and GEE labeling data by indicating N-linked glycans are necessary for antigen binding. Finally, mapping of the epitopes onto the antigen crystal structure revealed an approximate 90° relative spatial orientation, optimal for a noncompetitive binding ELISA. TEM data shows both linear and diamond antibody–antigen complexes with a similar binding orientation as predicted from the two footprinting-MS techniques. This study is the first of its kind to utilize multiple bottom-up footprinting-MS techniques and TEM visualization to characterize the monoclonal antibody-antigen binding interactions of critical reagents used in a quality control (QC) lot-release ELISA.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Protein structural characterization is an important aspect of biotherapeutic discovery and pharmaceutical development, primarily for the elucidation of structure-function relationships, investigations of mechanisms of action, and determining protein–protein interactions such as antibody–antigen binding interactions (epitope mapping). Epitope mapping is a critical aspect of biotherapeutic discovery and has historically been performed using alanine scanning, crystallography [1, 2], and nuclear magnetic resonance (NMR) [3]. While these are powerful tools that can provide residue-level and atomic resolution, their utility for epitope mapping can be limited by a target protein’s size or flexibility, relatively large sample requirements, or unpredictable structural changes associated with mutational analysis. Recent advancements in epitope mapping technologies include protein/peptide array [4, 5] and phage display [6], both of which can provide residue-specific information and/or enable high-throughput analysis; however, these methods are limited in their ability to detect conformational epitopes [7, 8] and their compatibility with glycoproteins.
Enabled by developments in instrumentation over the last decade, bottom-up mass spectrometry (MS) techniques have emerged as a valuable complementary approach that increase the ability to achieve peptide and residue-level information [9]. Hydrogen/deuterium exchange (HDX) is by far the most advanced of the bottom-up mass spectrometry-based techniques, and has emerged as the industry standard for epitope mapping of biotherapeutics approaches [10,11,12,13,14,15,16,17]. HDX has the advantage of providing nearly 100% protein sequence coverage, information on conformational epitopes, and has virtually no limit on the size of the protein(s) of interest [18,19,20]. However, challenges with this technology include back-exchange, deuterium scrambling, and rigorous data processing [21]. The advent of robotics for sample handling, online protein digestion procedures, and software improvements has enabled HDX to become a mainstream technology in the biotechnology industry. Many of the challenges described with HDX technology have been mitigated and now enable better reproducibility, high-throughput analysis, and faster data processing times. However, HDX has been proven to be difficult with heavily glycosylated proteins, and extensive efforts have been made to improve HDX analysis of glycoproteins with considerable success [22, 23].
Covalent labeling techniques coupled with mass spectrometry (footprinting-MS) have recently emerged as complementary bottom-up approaches for structural characterization and elucidation of protein–protein interactions [24, 25]. Recently, we have utilized both hydroxyl group footprinting-MS using fast photochemical oxidation of proteins (FPOP) and carboxyl group footprinting-MS with glycine ethyl ester labeling (GEE labeling), to separately characterize the binding interface of a Fab–antigen complex [26, 27]. This work was performed to demonstrate the utility of these technologies in the biotechnology industry sector.
Fast photochemical oxidation of proteins (FPOP) is a technique that utilizes a high power excimer laser to photo-dissociate hydrogen peroxide into hydroxyl radicals, facilitating the irreversible covalent labeling of protein residues as a measurement of solvent accessibility [28]. The solvent accessibility of a given region of a protein is based on the local microenvironment for the individual peptide/residue. Regions on the protein that are buried should be less solvent accessible than regions on the surface. Similarly, residues involved in protein–protein binding interactions will be less solvent accessible at the binding interface than in the unbound state. Using FPOP to measure the changes in solvent accessibility on an antigen in the bound versus unbound states has proven to be a reliable strategy for deciphering protein–protein binding interfaces [29,30,31,32]. However, determining if the differences in solvent accessibility are due to the specific binding interactions, or protein structural conformational changes that occur as a result of binding, is an inherent challenge as highlighted in this report.
Similar to FPOP, carboxyl group footprinting-MS using glycine ethyl ester (GEE) labeling is an irreversible covalent labeling technique that selectively labels carboxylic acids moieties (aspartic acid, glutamic acid, C-terminus) [33, 34]. Although hydroxyl radicals have been shown to react with all amino acids, residues containing the carboxylic acid moiety (aspartic acid, glutamic acid) are at least an order of magnitude less reactive than sulfur (methionine, cysteine) and aromatic containing residues (tryptophan, phenylalanine, tyrosine, histidine) [35]. Therefore, GEE labeling provides complementary data on residues that are limited by FPOP analysis. This technique has the added advantage of being able to monitor protein structural changes under experimental conditions difficult for other technologies such as the presence of lipid environments [33]. In addition, GEE labeling compares the rates of labeling of the antigen in the unbound versus bound states, enabling normalization in any global differences in solvent accessibility between an unbound and bound state [27, 34].
FPOP and GEE labeling provide unique and complementary information for determining peptides/residues involved in binding interfaces; however, demonstrated applications in which both methods have been required for structural elucidations are limited [36]. Here we describe the use of these technologies in combination to characterize the antibody–antigen interactions of a monoclonal antibody (mAb)-based sandwich enzyme-linked immunosorbant assay (ELISA) used for the detection and quantitation of a specific residual host cell protein, Phospholipase B-like 2 (PLBL2).
Therapeutic proteins are traditionally recombinantly expressed in hosts such as Chinese hamster ovary (CHO) cells [37]. During the fermentation process, host cell proteins (HCPs) are released into the cell culture fluid through natural secretion processes and from cell lysis. In order to ensure appropriate clearance of HCPs in the final product, extensive purification steps are usually required [38]. Unacceptable levels of HCPs in the final product pose a significant risk to patients, as they may lead to potential immunogenic effects [39]. Therefore, HCP levels are routinely monitored and quantitated using a variety of approaches, including ELISAs, Western blots, and proteomic mass spectrometry (reviewed in [40]), the results of which are typically required for lot-release of clinical and commercial biotherapeutics. Phospholipase B-like 2 (PLBL2) is a CHO-derived HCP previously identified to co-purify with several Genentech, Inc. clinical products [41] that required the development of a PLBL2-specific ELISA to measure residual PLBL2 in clinical material for lot-release testing. The PLBL2 sandwich ELISA utilizes two hybridoma-derived monoclonal antibodies (clones mAb1 and mAb2) with noncompetitive binding to PLBL2 and with roughly similar binding affinities. This assay is a quality control lot-release assay in which assay consistency and performance are of utmost importance.
CHO-derived PLBL2 is homologous to the previously identified lysosomal 66.3 kDa protein structurally defined as a member of the N-terminal nucleophile hydrolase superfamily [42]. The crystal structure for the mouse homolog indicates that this protein consists of two subunits with an αββα core fold, which herein are referred to as the N-terminal subunit (28 kDa) and C-terminal subunit (40 kDa). The protein has been shown to be highly glycosylated and displays significant heterogeneity in both O-linked and N-linked glycoforms, including varying levels of sialic acid and mannose-6-phosphate [43]. To investigate the potential impact of the glycan heterogeneity on performance of the immunoassay, we performed routine characterization studies using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot analysis. This analysis revealed that the detection monoclonal antibody (mAb1) binds to PLBL2 in the Western blot format, whereas the capture monoclonal antibody (mAb2) does not. These data suggested that mAb2 may bind to a conformational epitope [7]. We therefore sought to understand the nature of the binding interactions within the ELISA format to help ensure the consistency and performance of the critical reagents. Since our traditional methods of characterization were insufficient to elucidate the binding regions of the mAbs, we performed epitope mapping of the two PLBL2-mAb complexes.
This work describes the combination of FPOP and GEE labeling for determining epitopes on a highly glycosylated antigen. The FPOP data shows a significant suppression in labeling across the antigen in both complexes, which we attribute to conformational stability of the antigen. While these data were difficult to interpret in isolation, the putative epitopes identified spanned regions that contained N-linked glycans with proximal aspartic acid residues, prompting us to use GEE labeling for confirmatory studies. Combining the data from these technologies enabled us to confidently determine that regions containing N-linked glycans were involved in the interface of the antibody–antigen complexes. Subsequently we deglycosylated the intact antigen, resulting in distinct deglycosylation time-dependent loss in potency detected by the ELISA and verifying that N-linked glycans are involved in binding interactions. Mapping of the putative epitopes on the antigen crystal structure revealed that the two epitopes appear to be in a 90° relative spatial orientation, and in the optimal orientation for noncompetitive binding. Finally, we analyzed the PLBL2-mAb1-mAb2 mixture using negative stain transmission electron microscopy (TEM) [44], which revealed both a linear (2 mAbs + 1 antigen) and a diamond antibody-antigen (2 mAbs + 2 antigens) complex, both of which demonstrate the near 90° angle spatial binding position predicted from our FPOP data. This work provides the first data that couples multiple footprinting-MS techniques with TEM for epitope mapping of an antigen used in a monoclonal antibody sandwich ELISA. Combining these three techniques has enabled a comprehensive understanding of the antibody–antigen binding interactions of the critical reagents used in this QC lot-release ELISA.
Experimental
Chemicals and Materials
H2O2 (30%), trifluoroacetic acid (TFA), formic acid (FA), L-glutamine, L-methionine, cytochrome c, glycine ethyl ester (GEE), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), Trizma base, catalase, dithiothreitol (DTT), sodium iodoacetate (IAA), and all HPLC-grade solvents (H2O and acetonitrile, ACN) were obtained from Sigma-Aldrich (St. Louis, MO, USA) at the highest purity available.
Preparation of Samples
In brief, PLBL2 protein was transiently expressed in Chinese hamster ovaries (CHO) cells and purified for immunization in mice. PLBL2-specific mAbs were then produced using hybridoma technology for use in a sandwich ELISA. In this report, mAb2 is the capture antibody and mAb1 is the detection antibody used in the ELISA as previously described [41]. Antibody–antigen complexes were formed by mixing each mAb with the antigen at an excess molar ratio of 1:1.5 antigen:mAb to ensure complete binding of the antigen. Deglycosylation of PLBL2 was performed by incubation with either PNGaseF (P0704L, New England Biolabs, Ispwich, MA, USA) or EndoH (324717-200MIU, EMD Millipore, Burlington, MA, USA) using the manufacture’s recommendation conditions for 24 h and 72 h. Data for the relative % binding for the control samples is expressed in relation to the PLBL2 standard curve, and for the deglycosylated samples expressed in relation to the respective controls.
SDS Page Analysis and Western Blot Analysis
Samples were denatured with sodium dodecyl sulfate (SDS) (L3771, Sigma Aldrich, St. Lousi, MO, USA) and heated at 60 °C for 15 min with reducing agent dithiothreitol (DTT) (10 mM) (D0632, Sigma-Aldrich). Samples were then separated with a 4%–20% tris-glycine gel (1.0 mm × 20 well) using an Invitrogen xCell4 Surelock Midi-Cell electrophoresis system. The bands were visualized by SYPRO Ruby stain or immunoblotted. For the immunoblot method, proteins on the gels were transferred onto PVDF membranes: one gel to one membrane. Then, each PVDF membrane was incubated with mouse anti-PLBL2 antibody. The proteins were detected using a chemiluminescence system. Digitized images of the SYPRO® Ruby stained gels and immunoblots were acquired on the ChemiDoc XRS (Bio-Rad, Hercules, CA, USA).
FPOP Labeling Procedure
PLBL2 and the two PLBL2-specific antibody-antigen complexes (PLBL2-mAb1 and PLBL2-mAb2) were subjected to FPOP labeling as previously described [45]. Briefly, a 248 nm KrF excimer laser (GAM Laser Inc, Orlando, FL, USA) tuned to 50 mJ/pulse at 12 kV was focused with a convex lens (Edmund Optics, Barrington, NJ, USA) onto a 150 μm i.d. fused silica capillary and used to irradiate the sample. Two 1000 μL syringes, one containing H2O2 (HX0635, Sigma-Aldrich) and one containing a solution of protein and arginine (Arg) (A5006, Sigma-Aldrich) (protein+arginine), met at a micro-tee mixer located just before the point of laser irradiation. This allowed for mixing of the solutions from the two syringes and yielded a final concentration of 30 mM H2O2, 0.5 mg/mL total protein, and 30 mM arginine at the time of irradiation and hydroxyl radical generation. Both solutions were asymmetrically mixed with syringe pumps by using different capillary ID (75 μm for H2O2, 150 μm for (protein+Arg) and flow rates (450 μL/hr for H2O2, 900 μL/h for protein+Arg). The laser-pulse frequency was controlled by an external pulse generator (B&K Precision, Yorba Linda, CA, USA) and matched with the flow rate to allow for a 15% exclusion volume to ensure samples had only a single exposure to hydroxyl radicals. Labeled samples (150 μL) were collected in low protein-binding tubes containing 200 mM methionine (Met) (M9625, Sigma-Aldrich) and 3 μg of catalase (C40, Sigma-Aldrich) to quench residual H2O2 and hydroxyl radicals. FPOP analysis was performed using the previously described “equal-weight” strategy [26] in which the total amount of protein for each sample was kept constant (0.5 mg/mL) to minimize any differences in oxidative potential between the unbound and bound antigen solutions.
GEE Labeling Procedure
GEE labeling was performed in 25 mM TRIS, pH 7.5 labeling buffer at a final concentration of 688 mM GEE and 14.3 mM EDC (mol GEE:mol EDC = 48) as previously described [27]. Briefly, 200 μg of protein was used for each assay and the reactions were quenched after 0, 1.5, 3.9, 6.5, and 10 min reaction time with trifluoroacetic acid (TFA) at a final concentration of 0.6% v/v. The final volume for each assay was ~150 μL.
LC-MS/MS Analysis for FPOP and GEE Labeling
Samples were diluted with guanidine HCl (6 M) and DTT (10 mM, 45 °C for 10 min), s-carboxymethylated with 5 μL sodium iodoacetate (I2512, Sigma-Aldrich) (25 mM, at room temperature protected from light for 5 min), and quenched with DTT (50 mM, room temperature). The resulting samples were then desalted using NAP-5 columns (17-0853-02, GE Healthcare) with an elution volume of 800 μL PBS, digested with 5 μg trypsin (37 °C for 1 h), deglycosylated with 0.15 μg PNGaseF (37 °C for 30 min), and quenched with 30 μL of 100% formic acid (FA). Tryptic peptides (10 μg) were separated using a Waters H-Class UPLC with Waters Acquity UPLC CSH130 C-18 column (1.7 μm, 2.1 × 150 mm). Peptide separation occurred across a gradient from 100% solvent A (H2O, 0.1% FA) to 35% solvent B (ACN, 0.1% FA) over 42 min at a flow rate of 0.3 mL/min and column temperature of 77 °C. MS analysis was performed with a ThermoFisher QExactive operating in positive mode, performing MS2 scans on the top-10 most abundant peaks in data-dependent mode in the m/z range 350–2000 at a resolving power of 35,000.
Footprinting Mass Spectrometry
Tryptic peptide numbering for the PLBL2 antigen is based on the UnitPRot sequence ID# Q3TCN2. For FPOP and GEE labeling analysis, peak identification and quantitation of percent modification for each peptide were performed using Byonic and Byologic Footprint Software Suites, respectively (Protein Metrics, Inc.). For FPOP analysis, all samples were analyzed in triplicate. Each triplicate sample set had a corresponding “no laser” control, which was a sample run through the FPOP system without the laser pulsing, to account for background oxidation. The extracted ion chromatograms (XIC) for the oxidized peptide species, and the parent peptide were used to calculate the percent oxidation with the equation as previously described [26, 30, 32]. All of the observed charge states were included in the calculation for each peptide. Percent oxidation is presented as the average for triplicate runs after subtracting the “no laser” background oxidation control. Error bars represent the statistical analysis performed using a single sample t-test with a 95% confidence interval. Significant differences in % modification for a given peptide is determined by non-overlapping error bars comparing the unbound versus bound states.
For GEE labeling, a fixed modification was included for carboxymethyled cysteines (+58.0055 Da) and a variable modification was included for GEE mass shifts for GEE incorporated residues (+85.0528 Da). The GEE hydrolysis product (+57.0215 Da) was observed at trace levels and did not contribute significantly to the overall quantitation. The median change of the rate constant ratios was used to normalize to a value of 1 (division by the central tendency) as previously performed [27, 34], to compensate for non-biological variations between samples. Tryptic peptides with normalized rate constant ratio of 1 were considered to have no change in solvent accessibility. We designated significant differences in rates of labeling to be 50% or greater change from the value of 1 (e.g., ≤0.5 or ≥1.5), in which peptides with ratios ≥1.5 are considered more solvent exposed and those ≤0.5 are considered less solvent exposed in the bound state.
TEM Analysis
Negative stain transmission electron microscopy imaging and 2D class averaging were performed on a mixture of PLBL2, mAb1, and mAb2 by Nanoimaging Services. Sample preparation, imaging, analysis, and in silico renderings were performed as previously described [46].
Structural Mapping of Footprinting-MS Data
The mouse 66.3 kDa lysosomal homologue crystal structure (PDB:3FXB) was used as a surrogate for the CHO-derived PLBL2 protein for mapping of the FPOP and GEE labeling data. The PDB file was subjected to in silico N-linked glycosylation using GlyProt Freeware (http://www.glycosciences.de/modeling/glyprot/php/main.php).
Results and Discussion
Evaluation of mAb1 and mAb2 Binding Interaction with PLBL2
Monoclonal antibodies mAb1 and mAb2 were previously demonstrated to have noncompetitive binding in the sandwich ELISA format at relatively similar binding affinities for PLBL2 [41]. Western blot analysis indicated that mAb1 has a strong interaction with the full-length protein and binds weakly to the N-terminal subunit of the denatured and dissociated antigen (Figure 1). However, mAb2 did not bind the full-length protein or an antigen subunit under the reduced and denaturing conditions of the Western blot (Figure 1). These data imply that mAb2 may bind to a conformational epitope that requires the native, non-denatured higher order structure [7].
Characterization of the critical reagents used in the ELISA. (a) SDS-PAGE Gel analysis of PLBL2 shows full-length protein and the separation of the N-terminal and C-terminal subunits. (b) Western blot analysis indicates mAb1 binds to full-length PLBL2 and weakly to the N-terminus subunit, whereas mAb2 does not appear to bind PLBL2 under reduced/denaturing conditions. CHOP = CHO protein
FPOP/GEE Labeling Sequence Coverage and Glycopeptide Detection
The irreversible covalent labeling of FPOP and GEE labeling technologies enable the use of virtually any protease for bottom-up MS analysis. Trypsin is the protease of choice because of excellent reproducibility, efficiency, predictable cleavage sites, and ease of data analysis. We investigated the feasibility of using trypsin for footprinting-MS analysis and observed excellent sequence coverage (~90% total sequence coverage), post-deglycosylation of the peptides. In addition, our trypsin map of the native peptides identified the expected five N-linked glycosylation sites and over 20 putative O-linked glycosylation sites (Supplementary Figure S1). Previous literature has shown that the N-linked glycosylation sites are nearly fully occupied [43], but there is little information on the extent of O-linked glycosylation for PLBL2. Many of the O-linked glycans appear to be of low abundance in nature relative to the N-linked glycan occupancy (data not shown) and, therefore, were not considered in footprinting-MS analysis. All subsequent footprinting-MS analyses were performed on tryptic peptides that were treated with PNGaseF for N-linked glycan deglycosylation.
FPOP Analysis of PLBL2 in the Unbound State and Bound Complexes
The amount of hydroxyl radical labeling that occurs during FPOP analysis for a given region of a protein is determined by the solvent accessibility of the associated amino acid residues [29, 31]. Comparing regions of an antigen with reduced hydroxyl radical labeling in the unbound versus bound states can enable inference in the location of the epitope binding interface [29,30,31,32]. In this work, we subjected labeled samples to trypsin digestion to quantitate the amount of labeling (% modification) for each tryptic peptide in unbound PLBL2 and in bound complexes (PLBL2-mAb1 and PLBL2-mAb2). Using trypsin, we obtain >88% sequence coverage of FPOP labeled antigen peptides after deglycosylation of the tryptic digest using PNGaseF. Five tryptic peptides (tryptic peptide residues 80-99, 100-159, 212-237, 425-460, 514-539) were identified with the expected asparagine-to-aspartic acid modification that occurs from the N-linked deglycosylation by PNGaseF [47]. All of these tryptic peptides are located at positions that are consistent with known glycosylation sites when mapped onto the mouse-derived PLBL2 crystal structure [42] (subjected to in silico N-linked glycosylation).
Determining the binding interface from a protein–protein interaction using FPOP requires the observation of peptides/residues with reduction in solvent accessibility (measured by % modification) in the bound versus unbound state. Ideally, the reduction in % modification will be localized to specific regions of the protein for inference of the binding interface. Examining the full oxidative footprint of PLBL2 in the unbound and bound complexes, we observed a significant decrease in % modification in peptide labeling across the PLBL2 antigen in both complexes (Figure 2a). These data were difficult to interpret since the reduction in labeling was not localized to specific peptides in both complexes, and FPOP data cannot distinguish solvent accessibility changes that are due to the direct protein–protein interactions versus those due to subsequent conformational changes. We utilized our previous equal-weight strategy for FPOP samples analysis in which the total protein in solution is kept constant for the unbound and bound analyses [26]. Using this strategy, we previously showed that the protein has a negligible effect on the scavenging potential of FPOP samples at concentrations of ≤2 g/L, and therefore we do not think these broad reductions in peptide labeling are an artifact of the FPOP protocol.
We instead hypothesize that the antigen is in a more compact conformation in the antibody–antigen complexes [48], resulting in a significant change in the overall solvent accessibility of the protein. This “conformation stability” has been documented for antibody–antigen complexes using HDX as far back as the 1980s [49], and more recently using both HDX [50] and hydroxyl radical footprinting [51] with bound glycoproteins.
The FPOP data revealed that 23 of the 30 measured tryptic peptides exhibited suppression in reduction of solvent accessibility (% modification) in the PLBL2-mAb1 and PLBL2-mAb2 complexes (Figure 2a). These data on their own were nearly impossible to interpret for determining the binding interfaces for the two antibody-antigen complexes, requiring us to employ a novel strategy for elucidating the binding sites. First, 16 of these tryptic peptides showed similar suppression in both complexes (orange and yellow bars, respectively). To simplify our search for the epitopes, these peptides were mapped on to the mouse PLBL2 crystal structure and found to be located at the core of the protein (buried within the protein) (Figure 3). The changes in solvent accessibility for these peptides were considered to be due to conformational stability, and eliminated from the consideration of being associated with direct binding interactions. This left two distinct areas at the N-terminus and C-terminus with sequential peptides that exhibit suppression of % modification.
Secondly, we knew that the antibodies are noncompetitive and therefore have distinct binding regions. According to our Western blot data, mAb1 appears to bind weakly to the N-terminus of the antigen, and thus we attributed peptides with suppression in % modification in the bound state on the N-terminus (residues 57-65, 68-79, 80-99) to be associated with the mAb1 binding interface (blue bars). Finally, the remaining peptides located towards the C-terminus (residues 470-482, 486-513, 514-539) (red bars) were then attributed to mAb2 binding. Although this strategy was convincing, both mAbs appear to reduce the solvent accessibility of the N-terminus tryptic peptide containing residues 68-79, convoluting our putative identification. A complementary method was therefore required to strengthen confidence in the putative epitope identification.
GEE Labeling Analysis of PLBL2 in the Unbound State and Bound Complexes
Upon further examination of the FPOP data, we found that both of the putative epitopes on PLBL2 span peptides that contain N-linked glycans (residues 80-99 and 514-539, respectively) with neighboring aspartic acid residues. With this knowledge, we chose GEE labeling as a secondary footprinting-MS technique to further confirm the identity of epitopes. Trypsin digestion yielded >80% sequence coverage of the tryptic peptides containing carboxyl acid moieties in the antigen after deglycosylation of the peptides using PNGaseF. Interestingly, we observed significantly lower rates of GEE labeling for the antigen in the bound state compared with the unbound state (Supplementary Table S1), consistent with the FPOP data, which also demonstrated suppression of % modification in the bound state. Both data sets indicate that antigen binding results in more compact conformation, which we believe is due to conformational stability of the antigen. To compensate for the gross labeling differences, we utilized our normalization strategy in which the median change of the rate constant ratios was used to normalize to a value of 1 (division by the central tendency) [27, 34]. The normalization enabled us to obtain peptides on the antigen with significant decrease in GEE labeling in each antibody–antigen complex (Figure 2b). Consistent with the FPOP data, mAb1 appears to bind the N-terminus (residues 68-79, 80-99) and mAb2 appears to bind the C-terminus (residue 549-573). The N-terminus peptides identified by GEE labeling are consistent with those identified by FPOP; however, the C-terminus peptide(s) from the two data sets do not overlap. All three of the peptides identified by GEE labeling only contain a single GEE labeling site affording residue-level information. Aspartic acid (D) residues D72 and D82 are the labeling sites for the two N-terminus peptides, respectively, and D560 is the labeling site for the C-terminus peptide.
Footprinting-mass spectrometry data for the unbound PLBL2 and the PLBL2-mAb1 and PLBL2-mAb2 complexes. (a) FPOP: Comparison of the oxidative footprint of the PLBL2 tryptic peptides in the bound and unbound states. Peptides assigned to have conformational structural changes in the PLBL2-mAb1 and PLBL2-mAb2 complexes are shown in orange and yellow, respectively. Tryptic peptides involved in the putative binding regions for the PLBL2-mAb1 complex are shown in blue. Tryptic peptides involved in the putative binding regions for the PLBL2-mAb2 complex are shown in red. *Denotes significant changes in % modification of the bound versus unbound forms obtained from triplicate samples (p < 0.05). (b) GEE labeling: Comparison of the GEE rate of labeling ratio of bound versus unbound sates. Tryptic peptides involved in the putative binding regions for the PLBL2-mAb1 complex are shown in blue. Tryptic peptides involved in the putative binding regions for the PLBL2-mAb2 complex are shown in green. *Denotes significant changes in rate of GEE labeling of the bound versus unbound forms (>50% change rate). NL = not labeled
Structural Mapping of Footprinting-MS Data
Mapping the FPOP and GEE labeling data onto the mouse PLBL2 crystal structure enables us to visualize the binding regions for both mAbs (Figure 3). Both mAbs appear to bind regions containing N-linked glycans (shown as magenta sticks) on opposite sides of the PLBL2 antigen (Figure 3, left side). The FPOP and GEE labeling identify overlapping peptides (blue residues) of mAb1 binding site on the N-terminus of PLBL2. For the C-terminus epitope of mAb2, data from FPOP and GEE labeling (red and green residues, respectively) do not identify overlapping peptides; however, structural mapping demonstrates the proximity of D560 to the peptides identified via FPOP.
Footprinting-MS data mapped on to the mouse PLBL2 crystal structure (PDB:3FBX with in silico glycans added in magenta). Left structure: peptides putatively involved in the antibody-antigen binding regions for mAb1 (blue) and mAb2 (red and green). Right structure: peptides putatively exhibiting conformational structural changes upon mAb binding
As indicated above, mapping of the tryptic peptides that had similar changes in solvent accessibility (presumed to be conformational changes) demonstrated that most of the peptides were located toward the center of the molecule, between both of the binding regions (Figure 3, right side). Visualizing the footprinting-MS data on a representative 3D structure indicates that the epitopes appear to have an approximate 90° angle from each other, an optimal spatial position for the noncompetitive binding interactions of the sandwich ELISA.
Structural mapping also reveals a putative epitope for mAb2 that contains a high density of alpha helical secondary structure, which is consistent with its lack of antigen binding under reduced Western blot analysis. However, it is still unclear why mAb1 binds full length antigen and only weakly to the dissociated N-terminus subunit.
Effects of Deglycosylation on the Binding Affinity of PLBL2
Data from both footprinting-MS methods identified that the two putative epitopes appear to be associated with N-linked glycopeptides. To test the hypothesis that N-linked glycans contribute to the epitope binding regions, we deglycosylated the intact PLBL2 with PNGaseF and EndoH, and re-analyzed the deglycosylated antigen using the sandwich ELISA. Unfortunately, both PNGaseF and EndoH are similar in mass as the PLBL2 C-Terminus and N-Terminus, respectively, limiting the information that can be obtained from the dissociated antigen (Supplementary Figure S2). In addition, we found that the deglycosylation efficiency was limited for the intact antigen compared with the denatured antigen for both enzymes after 72 h of incubation (Supplementary Figure S2). These results were not unexpected because of the complexity of the antigen glycans (data not shown) [42]. Nonetheless, we observed a deglycosylation time-dependent loss in potency following treatment with both enzymes from 24 to 72 h (Figure 4), with no effect on the control sample (72 h with no deglycosylase). These data indicate that the epitopes involve N-linked glycan-specific interactions, supporting our FPOP and GEE labeling data.
Negative Stain Transmission Electron Microscopy (TEM) Imaging
After identifying the putative binding sites on either side of the antigen, we investigated the orientation of the antibody–antigen complex by visualization of negative stain TEM [44]. This method can provide relatively high-resolution images in nanometer scale for the static state structure found in a protein solution. The data from our TEM analysis provided a unique insight into the sandwich ELISA structural interactions on both the unbound mAbs and bound complexes (Figure 5a).
Negative stain transmission electron microscopy images for the unbound mAbs and antibody-antigen complexes. (a) Sample image at 110,000× magnification with 100 nm scale bar. Unbound mAb formations (B1 and B2) and antibody-antigen complexes (C1 and C2) are identified using square boxes. (b) Two distinct unbound mAb conformations were observed with differences in the Fab arm orientation. (1) Loop-shaped (planar Fab/Fc) orientation with possible interpretation of domain structure shown in blue. (2) Lobe-shaped (perpendicular Fab/Fc) orientation with possible interpretation of domain structure shown in red. (c) Two distinct antibody-antigen complexes were observed with mAb orientations similar to the unbound structures. (1) Linear complex and (2) diamond complex with possible interpretation of the mAb domain structures with the complexes (blue and red) and identification of the PLBL2 antigen (black circle)
Two distinct structural classifications were obtained for the unbound mAbs (Figure 5b). We observed a conical mAb three-ringed structure that appeared to have a planar orientation (denoted as looped-structure) (Figure 5b-1), and a structure with the Fab arms that appeared to be perpendicular to the Fc region (denoted as lobed-structure) (Figure 5b-2). In addition, two distinct structural classifications of the antibody–antigen complexes were observed: a linear complex (one antigen bound between 2 mAbs) (Figure 5c-1), and a diamond shaped complex (two antigens bound between two mAbs) (Figure 5c-2). Both antibodies have similar structures in the bound and unbound forms, indicating that the conformations of the unbound forms are retained in the complexes and not an artifact of the immobilization/staining process. Structural renderings are provided for the possible shapes of the unbound planar mAb (blue structure) and unbound perpendicular mAb (red structure). It is clear that the two mAbs appear to bind in a completely different orientation in both of the complexes. In addition, the angle of the dual binding event for both the linear and diamond complexes is consistent with the 90° angle spatial orientation predicted from the mapping of the footprinting-MS data onto the structure of the antigen.
Conclusion
In this work, we combined multiple footprinting-MS methods and TEM analysis to characterize and visualize the antibody-antigen interactions for a monoclonal sandwich ELISA used for QC lot-release testing. This extensive characterization was performed because of the unique questions surrounding glycan impact on PLBL2 antigen-antibody interactions, which are not typically examined by more traditional methods of reagent characterization. Our work provides the first-of-its-kind analysis combining bottom-up footprinting-MS and TEM visualization to elucidate the binding interactions of a ternary antibody-antigen complex.
We observed that much of the antigen showed suppression in solvent accessibility using FPOP when bound to either mAb, suggesting that conformational stability may be occurring upon binding. Glycoproteins are highly dynamic [52, 53] and the phenomenon of conformational stability has been documented [48, 49], but strategies for interpreting hydroxyl radical footprinting data for these circumstances have not been addressed. Here, we hypothesized that peptides with similar changes in solvent accessibility across both mAb1 and mAb2 complexes were the result of conformational stability in the antigen rather than direct binding events. These peptides mapped to the core of the antigen, supporting this hypothesis and simplifying interpretation of the remaining data (Figure 2 and Figure 3).
Combining these data with Western blot analysis enabled us to subsequently map the remaining data to the N-terminus and C-terminus (Figure 2, blue bars and red bars, respectively). Although this strategy was compelling for the identification of the two epitopes, we obtained confounding data for peptide 68-79, which required us to employ a confirmatory method. We identified the epitope regions spanned peptides containing both N-linked glycans, potentially limiting the technologies used for confirmatory studies. However, we observed that the putative epitopes appeared to involve aspartic acid residues, affording the use of GEE labeling as our complementary footprinting-MS technology. GEE labeling provided confirmatory data for the identification of the epitopes. Loss of potency in the ELISA after deglycosylation of the intact antigen confirmed that N-linked glycans are important for binding. Finally, mapping of the data from both footprinting-MS methods onto the mouse PLBL2 crystal structure provided the putative binding orientation of the two mAbs that was corroborated by our TEM analysis. In addition, TEM analysis provided insight into the different binding orientations of the two mAbs in both the linear and diamond ternary complexes.
In summary, the combination of FPOP, GEE labeling, and TEM enabled the identification of the putative binding epitopes for mAbs used in a sandwich ELISA. This integrated approach using multiple bottom-up footprinting-MS methods and TEM visualization for protein structural analysis is the first of its kind for examining monoclonal antibody-antigen interactions within a sandwich ELISA. Moreover, the use of high order structural techniques may be required to ensure consistency in critical reagents, when traditional characterization methods are not sufficient.
References
Weiss, G.A., Watanabe, C.K., Zhong, A., Goddard, A., Sidhu, S.S.: Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc. Natl. Acad. Sci. USA. 97, 8950–8954 (2000)
Muller, Y.A., Chen, Y., Christinger, H.W., Li, B., Cunningham, B.C., Lowman, H.B., de Vos, A.M.: VEGF and the Fab fragment of a humanized neutralizing antibody: crystal structure of the complex at 2.4 A resolution and mutational analysis of the interface. Structure (London, England : 1993). 6, 1153–1167 (1998)
Bardelli, M., Livoti, E., Simonelli, L., Pedotti, M., Moraes, A., Valente, A.P., Varani, L.: Epitope mapping by solution NMR spectroscopy. J. Mol. Recognit. 28, 393–400 (2015)
Katz, C., Levy-Beladev, L., Rotem-Bamberger, S., Rito, T., Rudiger, S.G., Friedler, A.: Studying protein-protein interactions using peptide arrays. Chem. Soc. Rev. 40, 2131–2145 (2011)
Hansen, C.S., Osterbye, T., Marcatili, P., Lund, O., Buus, S., Nielsen, M.: ArrayPitope: automated analysis of amino acid substitutions for peptide microarray-based antibody epitope mapping. PLoS One 12, e0168453 (2017)
Moreira, G., Fuhner, V., Hust, M.: Epitope mapping by phage display. Methods Mol. Biol. 1701, 497–518 (2018)
Forsstrom, B., Axnas, B.B., Rockberg, J., Danielsson, H., Bohlin, A., Uhlen, M.: Dissecting antibodies with regards to linear and conformational epitopes. PLoS One 10, e0121673 (2015)
Liang, Y., Guttman, M., Davenport, T.M., Hu, S.L., Lee, K.K.: Probing the impact of local structural dynamics of conformational epitopes on antibody recognition. Biochemistry. 55, 2197–2213 (2016)
Opuni, K.F., Al-Majdoub, M., Yefremova, Y., El-Kased, R.F., Koy, C., Glocker, M.O.: Mass spectrometric epitope mapping. Mass Spectrom Rev. (2016). https://doi.org/10.1002/mas.21516
Pradzinska, M., Behrendt, I., Astorga-Wells, J., Manoilov, A., Zubarev, R.A., Kolodziejczyk, A.S., Rodziewicz-Motowidlo, S., Czaplewska, P.: Application of amide hydrogen/deuterium exchange mass spectrometry for epitope mapping in human cystatin C. Amino Acids. 48, 2809–2820 (2016)
Baerga-Ortiz, A., Hughes, C.A., Mandell, J.G., Komives, E.A.: Epitope mapping of a monoclonal antibody against human thrombin by H/D-exchange mass spectrometry reveals selection of a diverse sequence in a highly conserved protein. Prot. Sci. 11, 1300–1308 (2002)
Huang, R.Y., Krystek, S.R., Jr., Felix, N., Graziano, R.F., Srinivasan, M., Pashine, A., Chen, G.: Hydrogen/deuterium exchange mass spectrometry and computational modeling reveal a discontinuous epitope of an antibody/TL1A Interaction. MAbs 1–9 (2017)
Li, J., Wei, H., Krystek Jr., S.R., Bond, D., Brender, T.M., Cohen, D., Feiner, J., Hamacher, N., Harshman, J., Huang, R.Y., Julien, S.H., Lin, Z., Moore, K., Mueller, L., Noriega, C., Sejwal, P., Sheppard, P., Stevens, B., Chen, G., Tymiak, A.A., Gross, M.L., Schneeweis, L.A.: Mapping the energetic epitope of an antibody/interleukin-23 interaction with hydrogen/deuterium exchange, fast photochemical oxidation of proteins mass spectrometry, and alanine shave mutagenesis. Anal. Chemi. 89, 2250–2258 (2017)
Zhang, Q., Willison, L.N., Tripathi, P., Sathe, S.K., Roux, K.H., Emmett, M.R., Blakney, G.T., Zhang, H.M., Marshall, A.G.: Epitope mapping of a 95 kDa antigen in complex with antibody by solution-phase amide backbone hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 83, 7129–7136 (2011)
Lim, X.X., Chandramohan, A., Lim, X.E., Crowe Jr., J.E., Lok, S.M., Anand, G.S.: Epitope and paratope mapping reveals temperature-dependent alterations in the Dengue-antibody interface. Structure (London, England: 1993). 25, 1391–1402 e1393 (2017)
Terral, G., Champion, T., Debaene, F., Colas, O., Bourguet, M., Wagner-Rousset, E., Corvaia, N., Beck, A., Cianferani, S.: Epitope characterization of anti-JAM-A antibodies using orthogonal mass spectrometry and surface plasmon resonance approaches. MAbs. 9, 1317–1326 (2017)
Domina, M., Lanza Cariccio, V., Benfatto, S., Venza, M., Venza, I., Donnarumma, D., Bartolini, E., Borgogni, E., Bruttini, M., Santini, L., Midiri, A., Galbo, R., Romeo, L., Patane, F., Biondo, C., Norais, N., Masignani, V., Teti, G., Felici, F., Beninati, C.: Epitope mapping of a monoclonal antibody directed against neisserial heparin binding antigen using next generation sequencing of antigen-specific libraries. PLoS One. 11, e0160702 (2016)
Mayne, L.: Hydrogen exchange mass spectrometry. Methods Enzymol. 566, 335–356 (2016)
Huang, R.Y., Chen, G.: Higher order structure characterization of protein therapeutics by hydrogen/deuterium exchange mass spectrometry. Anal. Bioanal. Chem. 406, 6541–6558 (2014)
Lee, J.J., Park, Y.S., Lee, K.J.: Hydrogen-deuterium exchange mass spectrometry for determining protein structural changes in drug discovery. Arch. Pharm. Res. 38, 1737–1745 (2015)
Iacob, R.E., Engen, J.R.: Hydrogen exchange mass spectrometry: are we out of the quicksand? J. Am. Soc. Mass Spectrom. 23, 1003–1010 (2012)
Guttman, M., Scian, M., Lee, K.K.: Tracking hydrogen/deuterium exchange at glycan sites in glycoproteins by mass spectrometry. Anal. Chem. 83, 7492–7499 (2011)
Jensen, P.F., Comamala, G., Trelle, M.B., Madsen, J.B., Jorgensen, T.J., Rand, K.D.: Removal of N-linked glycosylations at acidic pH by PNGase A facilitates hydrogen/deuterium exchange mass spectrometry analysis of N-linked glycoproteins. Anal. Chem. 88, 12479–12488 (2016)
Mendoza, V.L., Vachet, R.W.: Probing protein structure by amino acid-specific covalent labeling and mass spectrometry. Mass Spectrom. Rev. 28, 785–815 (2009)
Wang, L., Chance, M.R.: Protein footprinting comes of age: mass spectrometry for biophysical structure assessment. Mol. Cell. Proteom. 16, 706–716 (2017)
Zhang, Y., Wecksler, A.T., Molina, P., Deperalta, G., Gross, M.L.: Mapping the binding interface of VEGF and a monoclonal antibody Fab-1 fragment with fast photochemical oxidation of proteins (FPOP) and mass spectrometry. J. Am. Soc. Mass Spectrom. 28(5), 850–858 (2017)
Wecksler, A.T., Kalo, M.S., Deperalta, G.: Mapping of Fab-1:VEGF interface using carboxyl group footprinting mass spectrometry. J. Am. Soc. Mass Spectrom. 26, 2077–2080 (2015)
Chen, J., Rempel, D.L., Gau, B.C., Gross, M.L.: Fast photochemical oxidation of proteins and mass spectrometry follow submillisecond protein folding at the amino-acid level. J. Am. Chem. Soc. 134, 18724–18731 (2012)
Jones, L.M., J, B.S., J, A.C., Gross, M.L.: Fast photochemical oxidation of proteins for epitope mapping. Anal. Chem. 83, 7657–7661 (2011)
Xu, G., Chance, M.R.: Radiolytic modification of acidic amino acid residues in peptides: probes for examining protein-protein interactions. Anal. Chem. 76, 1213–1221 (2004)
Loizos, N.: Mapping protein-ligand interactions by hydroxyl-radical protein footprinting. Methods Mol. Biol. 261, 199–210 (2004)
Yan, Y., Chen, G., Wei, H., Huang, R.Y., Mo, J., Rempel, D.L., Tymiak, A.A., Gross, M.L.: Fast photochemical oxidation of proteins (FPOP) maps the epitope of EGFR binding to adnectin. J. Am. Soc. Mass Spectrom. 25, 2084–2092 (2014)
Collier, T.S., Diraviyam, K., Monsey, J., Shen, W., Sept, D., Bose, R.: Carboxyl group footprinting mass spectrometry and molecular dynamics identify key interactions in the HER2-HER3 receptor tyrosine kinase interface. J. Biol. Chem. 288, 25254–25264 (2013)
Kaur, P., Tomechko, S.E., Kiselar, J., Shi, W., Deperalta, G., Wecksler, A.T., Gokulrangan, G., Ling, V., Chance, M.R.: Characterizing monoclonal antibody structure by carboxyl group footprinting. MAbs 7, 540–552 (2015)
Xu, G., Chance, M.R.: Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem. Rev. 107, 3514–3543 (2007)
Li, K.S., Chen, G., Mo, J., Huang, R.Y., Deyanova, E.G., Beno, B.R., O'Neil, S., Tymiak, A.A., Gross, M.L.: Orthogonal mass spectrometry-based footprinting for epitope mapping and structural characterization: the IL-6 receptor upon binding of protein therapeutics. Anal. Chem. 89(14), 7742–7749 (2017)
Dingermann, T.: Recombinant therapeutic proteins: production platforms and challenges. Biotechnol. J. 3, 90–97 (2008)
Bracewell, D.G., Francis, R., Smales, C.M.: The future of host cell protein (HCP) identification during process development and manufacturing linked to a risk-based management for their control. Biotechnol. Bioeng. 112, 1727–1737 (2015)
de Zafra, C.L., Quarmby, V., Francissen, K., Vanderlaan, M., Zhu-Shimoni, J.: Host cell proteins in biotechnology-derived products: a risk assessment framework. Biotechnol. Bioeng. 112, 2284–2291 (2015)
Zhu-Shimoni, J., Yu, C., Nishihara, J., Wong, R.M., Gunawan, F., Lin, M., Krawitz, D., Liu, P., Sandoval, W., Vanderlaan, M.: Host cell protein testing by ELISAs and the use of orthogonal methods. Biotechnol. Bioeng. 111, 2367–2379 (2014)
Vanderlaan, M., Sandoval, W., Liu, P., Nishihara, J., Tsui, G., Lin, M., Gunawan, F., Parker, S., Wong, R.M., Low, J., Wang, X., Yang, J., Veeravalli, K., McKay, P., Yu, C., O’Connell, L., Tran, B., Vij, R., Fong, C., Francissen, K., Zhu-Shimoni, J., Quarmby, V., Krawitz, D.: Hamster phospholipase B-like 2 (PLBL2): a host-cell protein impurity in therapeutic monoclonal antibodies derived from Chinese hamster ovary cells. BioProcess Int. 13, 18–55 (2015)
Lakomek, K., Dickmanns, A., Kettwig, M., Urlaub, H., Ficner, R., Lubke, T.: Initial insight into the function of the lysosomal 66.3 kDa protein from mouse by means of X-ray crystallography. BMC Struct. Biol. 9, 56 (2009)
Deuschl, F., Kollmann, K., von Figura, K., Lubke, T.: Molecular characterization of the hypothetical 66.3-kDa protein in mouse: lysosomal targeting, glycosylation, processing and tissue distribution. FEBS Lett. 580, 5747–5752 (2006)
De Carlo, S., Harris, J.R.: Negative staining and cryo-negative staining of macromolecules and viruses for TEM. Micron. 42, 117–131 (2011)
Zhang, Y., Rempel, D.L., Zhang, H., Gross, M.L.: An improved fast photochemical oxidation of proteins (FPOP) platform for protein therapeutics. J. Am. Soc. Mass Spectrom. 26, 526–529 (2015)
Sung, J.J., Pardeshi, N.N., Mulder, A.M., Mulligan, S.K., Quispe, J., On, K., Carragher, B., Potter, C.S., Carpenter, J.F., Schneemann, A.: Transmission electron microscopy as an orthogonal method to characterize protein aggregates. J. Pharm. Sci. 104, 750–759 (2015)
Tarentino, A.L., Trimble, R.B., Plummer Jr., T.H.: Enzymatic approaches for studying the structure, synthesis, and processing of glycoproteins. Methods Cell. Biol. 32, 111–139 (1989)
Roguin, L.P., Retegui, L.A.: Monoclonal antibodies inducing conformational changes on the antigen molecule. Scand. J. Immunol. 58, 387–394 (2003)
Zavodszky, P., Jaton, J.C., Venyaminov, S.Y., Medgyesi, G.A.: Increase of conformational stability of homogeneous rabbit immunoglobulin G after hapten binding. Mol. Immunol. 18, 39–46 (1981)
Guttman, M., Kahn, M., Garcia, N.K., Hu, S.L., Lee, K.K.: Solution structure, conformational dynamics, and CD4-induced activation in full-length, glycosylated, monomeric HIV gp120. J. Virol. 86, 8750–8764 (2012)
Li, X., Grant, O.C., Ito, K., Wallace, A., Wang, S., Zhao, P., Wells, L., Lu, S., Woods, R.J., Sharp, J.S.: Structural analysis of the glycosylated intact HIV-1 gp120-b12 antibody complex using hydroxyl radical protein footprinting. Biochemistry. 56, 957–970 (2017)
Mallajosyula, S.S., Jo, S., Im, W., MacKerell Jr., A.D.: Molecular dynamics simulations of glycoproteins using CHARMM. Methods Mol. Biol. 1273, 407–429 (2015)
Wormald, M.R., Dwek, R.A.: Glycoproteins: glycan presentation and protein-fold stability. Structure (London, England: 1993). 7, R155–R160 (1999)
Acknowledgements
The authors thank Natalie K. Garcia, Shelley Elvington, Matt S. Kalo, and Heidi Zhang for thoughtful review of this manuscript. The authors thank Tomasz Baginski for providing glycan expertise, Kathy Fourre, Kim Reyes and Julie Nishihara for Western blot and SDS-PAGE analysis.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 706 kb)
Rights and permissions
About this article
Cite this article
Lin, M., Krawitz, D., Callahan, M.D. et al. Characterization of ELISA Antibody-Antigen Interaction using Footprinting-Mass Spectrometry and Negative Staining Transmission Electron Microscopy. J. Am. Soc. Mass Spectrom. 29, 961–971 (2018). https://doi.org/10.1007/s13361-017-1883-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-017-1883-9
Keywords
- Fast photochemical oxidation of proteins (FPOP)
- Hydroxyl-radical footprinting-mass spectrometry
- Carboxyl-group footprinting-mass spectrometry
- GEE labeling
- Negative staining transmission electron microscopy (TEM)
- Phospholipase B-like 2 (PLBL2)
- Enzyme-linked sandwich immunosorbant assay (ELISA)
- Conformational stability