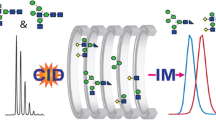
Abstract
Differential mobility spectrometry (DMS) has been employed to separate isomeric species in several studies. Under the right conditions, factors such as separation voltage, temperature, the presence of chemical modifiers, and residence time can combine to provide unique signal channels for isomeric species. In this study, we examined a set of glycopeptide isomers, MUC5AC-3 and MUC5AC-13, which bear an N-acetyl-galactosamine (GalNAc) group on either threonine-3 or threonine-13. When analyzed as a mixture, the resulting MS and MS/MS spectra yield fragmentation patterns that cannot discern these convolved species. However, when DMS is implemented during the analysis of this mixture, two features emerge in the DMS ionogram representing the two glycopeptide isomers. In addition, by locking in DMS parameters at each feature, we could observe several low intensity CID fragments that contain the GalNAc functionality-specific amino acid residues – identifying the DMS separation of each isomer without standards. Besides conventional CID MS/MS, we also implemented electron-capture dissociation (ECD) after DMS separation, and clearly resolved both isomers with this fragmentation method, as well. The electron energy used in these ECD experiments could be tuned to obtain maximum sequence coverage for these glycopeptides; this was critical as these ions were present as doubly protonated species, which are much more difficult to fragment efficiently via electron-transfer dissociation (ETD). Overall, the combination of DMS with electron- or collision-based MS/MS methods provided enhanced separation and sequence coverage for these glycopeptide isomers.

ᅟ
Similar content being viewed by others
Introduction
The glycosylation of proteins is a fairly common post-translational modification [1] that affects such properties as cell signaling and solubility. In recent years, mass spectrometry (MS) has proven to be an essential tool in the characterization of glycoproteins and glycopeptides [2] based upon the overall sensitivity and selectivity of the technique. In terms of the latter quality, the fragmentation methods available on modern MS systems can provide peptide sequence as well as glycan sequence information depending upon the fragmentation method chosen [3]. For example, some structural information related to glycosylation sites can be retained using traditional collision-induced dissociation (CID) [3, 4] albeit at low relative abundance to other fragment ions. However, alternative MS/MS methods to CID are generally implemented to retain otherwise labile glycan groups on peptide fragment ions—electron-capture dissociation (ECD) [5, 6] and electron-transfer dissociation (ETD) [7, 8]. These methods have also been blended in elegant ways to provide CID and ECD/ETD complementary information in back-to-back scans for a given precursor glycopeptide ion [9, 10].
Although powerful, even these modern MS and MS/MS methodologies can face challenges, especially when convolved mixtures of isomeric glycopeptides are analyzed. When little separation of such isomers is provided by traditional means, such as reversed-phase liquid chromatography (RPLC), the resulting CID or ECD/ETD analyses will yield convoluted fragmentation spectra, thereby limiting the accuracy with which glycosylation sites can be identified. Hence, additional orthogonal methods of separation are essential to providing more complete characterization of such glycopeptides. Among these methods is high-field asymmetric waveform ion mobility (FAIMS), also known as differential mobility spectrometry (DMS) [11–14]. As implemented in this study, the DMS cell consists of two parallel planar electrodes (30 × 10 mm) separated by a 1-mm gap. Ions generated by an electrospray ionization (ESI) source are carried by the transport gas (curtain gas) into this cell while an asymmetric rf waveform (i.e., separation voltage or SV at 3 MHz) is applied in the transverse direction to the gas flow. This causes ions to oscillate rapidly toward one electrode or the other, depending upon the mobility behavior of the ion. In order to direct ions toward the on-axis sampling orifice of the MS, a second DC potential is applied to the electrodes (i.e., compensation voltage or CV).
In recent years, FAIMS/DMS has been used to separate isomers of many forms, including structural isomers [15–17], sequence variant peptides [18], and tautomers [19–21]. Here, we describe the DMS separation of a set of glycopeptide isomers to enhance their characterization by either collision- or electron-based tandem mass spectrometry methods. We chose a model system earlier employed by Creese and Cooper [22], the glycoprotein MUCIN 5AC, which bears an O-linked N-acetylgalactosamine (GalNAc) group at either the threonine-3 or threonine-13 amino acid residue. Although not separable by RPLC, Creese and Cooper utilized FAIMS to provide a small margin of separation prior to analysis by ETD. However, the separation power of their cylindrical FAIMS device was only moderate, and their ETD fragmentation was hampered by the inefficiencies inherent with dissociating lower charge state precursor ions with this technique [23, 24]. With our DMS-MS instrumentation, we make use of chemical modifiers [25, 26] to enhance the degree of separation of these two isomers prior to interrogation either by CID or by efficient ECD, performed using a unique ToF-MS-based cell [27].
Methods
Sample Preparation
The two model isomers employed were 16-amino acid glycopeptides from the Mucin 5AC protein: MUC5AC-3 and MUC5AC-13. The amino acid sequence for both peptides was identical (GTTPSPVPTTSTTSAP), with the two isomers differing only in the threonine (T) to which the GalNAc (*) functionality was linked: MUC5AC-3 = GTT*PSPVPTTSTTSAP and MUC5AC-13 = GTTPSPVPTTSTT*SAP). These glycopeptides were purchased from Anaspec (Fremont, CA, USA) and were used without further purification. The working ESI solutions of these glycopeptides (analyzed either individually or in a 1:1 mixture) were prepared to 1 μM with acetonitrile and water (50/50, v/v) containing 0.1% formic acid.
DMS-MS Instrumentation
Experiments were performed using either a modified hybrid quadrupole-linear ion trap mass spectrometer (qLIT) or a modified quadrupole time-of-flight (q-ToF) mass spectrometer (SCIEX, Concord, ON, Canada). Both instruments are equipped with identical differential mobility spectrometer (DMS) hardware (SCIEX), the fundamental properties of which have been described elsewhere [11, 13]. The DMS cell is mounted between each mass spectrometer’s sampling orifice and a Turbo V ion source (Ion Spray ESI voltage of +5000 V) (Figure 1, Q-ToF MS configuration). The temperature of the DMS cell was maintained at either 225 °C (Q-ToF experiments) or 150 °C (qLIT), with nitrogen curtain gas operated at 25 psi. Resolving gas (nitrogen) was also employed at 25 psi to enhance the separation of the isomers. Chemical modifier (acetonitrile) was added into the curtain gas flow at 1.5% (mole ratio).
For one of the DMS-based experiments conducted with the qLIT MS, we collected data to create so-called “dispersion plots” [27] that reveal the type of DMS behavior ions exhibit [12]. In these experiments, SV was stepped from 0 to 3500 V (in 500-V increments), and at each of these SV steps, CV was scanned from –10 V to +10 V. At each 0.25-V increment of CV, an MS/MS spectrum, the [M + 2H]2+ ion for the glycopeptide isomers (m/z 853) was recorded using a collision energy of 50 eV (lab frame). The resulting plot reveals the optimal CV at which an ion is transmitted through the DMS cell, and the curvature of the SV/CV plot can describe the behavior an ion exhibits within the DMS. In these experiments, acetonitrile was employed as the chemical modifier.
For other DMS experiments, the SV for the DMS cell was set at 3500 V while the CV was ramped between –10.0 and +10.0V. A resolving (throttle) gas pressure of 25 psi was also employed to improve resolution of the isomeric species [14]; the presence of this resolving gas increases the residence time of the ions within the DMS cell. After determination of the optimal CV values for the DMS-separated glycopeptide isomers, CV values were locked in while either ECD or CID mass spectra were acquired for each isomer. For the Q-ToF MS system, at each 0.10-V step in CV, a ToF-MS (or CID or ECD) mass spectrum (m/z 100–2000, resolving power ~30,000) was recorded. For the MS/MS spectra, the doubly protonated glycopeptides were chosen as the precursor ions (m/z 853) (vide infra). All data were processed using a research-grade version of PeakView (SCIEX).
The Q-ToF MS is equipped with an ECD cell installed between lens electrode IQ2 and quadrupole q2 (Figure 1) [28, 29]. As such, this instrument can collect ECD spectra of precursor ions isolated by Q1 or it can collect conventional beam-type CID fragmentation spectra by accelerating Q1-isolated precursor ions through the ECD cell and into q2. For performing ECD, electron energies ranging from 0.1 to 20.0 V (or greater) are available with this cell; here, an optimal setting of 5.0 eV was employed for these experiments. The CID experiments utilized a lab-frame energy of 35 eV. A more specific discussion of the ECD cell’s design and operation (including additional components and their respective manufacturers) is given elsewhere [28].
Results and Discussion
Developing Dispersion Plots Helps Evaluate the DMS Separation of Isomers
The creation of a “dispersion plot” [27] can be very useful for evaluating the DMS’s ability to separate isomers and to describe the behaviors of these ions that lead to their separation. As mentioned earlier, the SV applied to the DMS was stepped from 0 to 3500 V with CV scans from –10 V to +10 V performed at every 500-V SV step. The same experiment was conducted through these SV and CV scans – MS/MS of the doubly protonated MUCIN glycopeptides of m/z 853; this charge state was chosen for MS/MS given its vast relative abundance relative to the +1 and +3 charge states (vide infra). As displayed in Figure 2a, the CV scans recorded at progressively greater SV values show an increase in intensity up to SV = 2000 V, and then proceed to become smaller at progressively higher SV values [30]. No separation of the two glycopeptide isomers (present in a 1:1 ratio in the ESI solution) is evident until, at SV = 3000 V, a broadening of the ionogram (Figure 2, light green trace) begins to reveal features within the signal, culminating with the ionogram recorded at SV = 3500 and the appearance of two distinct peaks. As with many isomeric species (i.e., containing almost identical structures), the dispersion plot (SV versus CV) for these two glycopeptides are identical until the SV = 3500 mark (top of Figure 2b), where the two traces begin to diverge at slightly different rates.
Separation of Isomeric Glycopeptides by DMS Can be Charge State-Dependent
The DMS systems employed in this study have several variables at their disposal that allow for the separation of isomeric species. In this case, the separation of the isomeric glycopeptides MUC5AC-3 and MUC5AC-13 required the use of high SV (3500 V) combined with added chemical modifier (acetonitrile) and resolving gas (25 psi). As depicted in Figure 3a, we could obtain separation of these isomers in CV space in their doubly protonated forms – the charge state present in the greatest relative abundance (Figure 3b, Figure S1). An earlier FAIMS (i.e., cylindrical DMS) study of these same glycopeptide isomers also targeted the doubly protonated molecules (presumably for the same reason as our study), and used a mixture of carrier gases (He and N2) [31, 32] to separate the isomers, albeit with somewhat lower resolution [22]. However, while the full-width half maximum peak widths of the individual glycopeptide isomers in the DMS ionograms (1.1–1.4 V) are indeed much narrower than the FAIMS-based data (~6 V), we also observed some tailing on the negative CV side of the MUC5AC-3 signal (Figure 3a). This may be indicative of multiple structures present in that ion population (vide infra). The DMS’s ability to probe ion structure via the interactions of chemical modifier molecules provides such possibilities.
(a) Overlaid ionograms for three analyzed samples (all analytes present at 1 μM each): (1) MUC5AC-13 (pink), (2) MUC5AC-3 (red), and (3) an equimolar mixture of both glycopeptide isomers; (b) overlaid ionograms obtained during the analysis of the equimolar solution of glycopeptide isomers, releaving the impact of the glycopeptides’ charge states on the DMS’s ability to separate these species. For reference, an inset depicts the normalized/relative intensities of each charge state. N.B. The DMS temperature for these experiments conducted with the Q-ToF MS was 225 °C while the qLIT DMS was held at 150 °C. This accounts for the observed differences in CV shift between Figures 2 and 3, even though the separation quailty for the isomers is nearly equivalent
Interestingly, while the DMS settings used in this study could be used to separate the doubly protonated glycopeptide isomers, the two other charge states visible in the ToF mass spectra (+1 and +3) were inseparable under the same DMS conditions (Figure 3b). However, the peak width (in CV space) for the +1 charge state (~2.5 V) was almost double that of the +3 charge state (~1.1 V), suggesting that the glycopeptide isomers present as +1 charge state ions were close to being separated by the DMS. These facts point to a role of the sites of protonation and the charge states as critical factors in the DMS separation.
The MUC5AC glycopeptides are not considered “basic peptides” as they are non-tryptic peptides containing no lysine or arginine residues. As such, the sites of protonation other than the peptides’ N-termini are not as obvious. In fact, another study of similar glycopeptides [33] suggested many equally possible sites for the location of the second proton, each having nearly identical energies. How could these sites of protonation influence the DMS separation (or lack thereof) for the glycopeptides studied here? For the case of the separated doubly protonated species, the more negative CV shift by the MUC5AC-13 isomer suggests that it binds more strongly with the acetonitrile vapor added to the DMS cell than does the MUC5AC-3 isomer [34–36]. When considering the structural variation between isomers that could cause such a CV shift, the presence of possible intramolecular hydrogen bonds (IMHBs) in the MUC5AC-3 isomer that internally solvate a site of protonation could be proposed (vide infra). Ions that exhibit tendencies to form such IMHBs display more positive CV shifts than isomers without IMHBs [37]. The presence of IMHBs establishes a competition between an intramolecular and an intermolecular hydrogen bond with any acetonitrile molecules in the DMS cell; hence, the MUC5AC-13 isomer exhibits a more negative CV shift than the MUC5AC-3 isomer.
The doubly protonated isomers may have their sites of protonation at the N-terminus and at one of the four proline residues along the peptide backbone. This could establish a possible site for an IMHB in the MUC5AC-3 isomer between the GalNAc moiety and the protonated N-terminus, as well as with a protonated proline. By comparison, the MUC5AC-13 isomer would not exhibit the same proximity between the site of the GalNAc functionality and the protonated N-terminus, yielding a more accessible positive charge for binding to acetonitrile chemical modifier molecules (and a concomitantly more negative CV shift). A similar proposal could be made for the wide yet still convolved ionogram of the singly protonated MUC5AC glycopeptides. Here, a minor structural difference, possibly an IMHB in the MUC5AC-3 isomer between the protonated N-terminus and the GalNAc group, could also establish a difference in the binding interactions of the two isomers with acetonitrile molecules. As for the glycopeptide isomers existing with a +3 charge state, the addition of the third proton could establish a charge site easily accessible to acetonitrile molecules for both isomers, essentially leveling out any differences in structure between the two species. Given these results, the influence of charge state on the separation of peptide isomers is now under further investigation in our lab.
DMS-Separated Glycopeptide Isomers Yield Informative CID and ECD Fragmentation Patterns
Ultimately, the separation of these two glycopeptide isomers should provide a route to obtaining improved tandem mass spectra for these species, providing better identification and sequence coverage. With this in mind, we next focused on the quality of the CID and ECD spectra produced from the DMS-separated isomers. Comparing these data with the respective spectra collected from the “pure” isomers would demonstrate the DMS’ ability to provide isomeric separation and quality fragmentation spectra for individual isomers with a mixture.
As is expected when CID is performed on glycopeptide ions, the vast majority of the b and y fragment ions for the two peptides were identical for both isomers. The labile glycan group, once lost, leaves behind two identical peptide structures that provide few distinguishing features in their CID spectra. However, there were actually two informative CID fragment ions, the signal-to-noise of which was enhanced after DMS separation. As displayed in Figure 4, each of the two DMS-separated isomers transmitted at CV = –1.1V (MUC5AC-13) and at CV = +0.4V (MUC5AC-3) yielded unique weak, yet diagnostic, peptide fragment ions that retained the GalNAc modification on the fragmented peptide backbone. For MUC5AC-3, b3 + GalNAc was observed at m/z 463.2, while for MUC5AC-13, y9 + GalNAc was detected at m/z 1065.5. While generally thought of as very labile, some smaller glycan modifications can be retained upon CID and can be used as diagnostic features to localize glycosylation sites [3]. However, in the presence of other isomeric glycopeptides, such diagnostic peaks could become diluted without the separation afforded by the DMS.
A comparison of the CID mass spectra for the DMS-separated MUC5AC-13 (pink, top, CV = –1.1 V) and MUC5AC-3 (red, bottom, CV= +0.4 V). Although not immediately obvious from viewing the very similar CID fragmentation patterns in the full-scan mass spectrum (a), two minor diagnostic fragment ions are apparent upon closer inspection: (b) the b3 ion of MUC5AC-3 isomer at m/z 463.20 and (c) the y9 ion of MUC5AC-13 at m/z 1065.49
The ECD spectra collected from the DMS-separated MUC5AC-3 and MUC5AC-13 isomers were even more informative than the CID spectra. This was expected as previous studies involving either the ECD [6] or ETD [9, 10, 22] of glycopeptides have demonstrated that these dissociation techniques provide peptide backbone fragmentation with retention of labile glycan functionalities. As shown in Figure 5, the ECD fragmentation patterns of the two glycopeptide isomers reveal the retention of the GalNAc modification on either threonine-3 (for MUC5AC-3) or threonine-13 (for MUC5AC-13). These clear distinctions are based, in part, by our instrument’s ability to perform “hot ECD” (i.e., using electron energies in excess of 3 eV) [38, 39] to generate fragment ions efficiently for these glycopeptides. This capability to tune electron energies makes ECD more useful than ETD, especially for doubly charged precursors [23], given that the energetics of those dissociation reactions are dictated by the specific chemical properties of the individual reagent ion employed [40], the reaction kinetics are hampered by the relatively small total number of charges involved in this ion/ion reaction [24], and compensating for these slow kinetics by using “supplemental activation” with ETD often results in the loss of the glycan moiety via that activation method [23].
(a) Full-scan ECD mass spectra for the two DMS-separated glycopeptide isomers: MUC5AC-13 (top, pink) and MUC5AC-3 (bottom, red); (b)annotated ECD mass spectra for MUC5AC-13 (top, pink) and MUC5AC-3 (bottom, red) displaying the sequence coverage and glycosylation sites on each peptide (fragments characteristic of the GalNAc location indicated in colored boxes). Note the "25x" and "15x" labels at the top of the spectrum in (a), indictaing the magnification of the ion intensities within the regions marked by arrows
Conclusions
By combining DMS with both ECD and CID fragmentation, we obtained fragmentation spectra of qualities high enough to discern two isomeric glycopeptides, differing only in the location of the GalNAc functionality. While the differentiation and level of information provided by the ECD spectra are obvious, even the DMS-separated CID spectra revealed that these two glycopeptide isomers could be distinguished based upon two low-abundance fragment ions provided by this conventional method. Clearly, in situations where larger, more branched glycoforms can be found as modifications, ECD would be the obvious choice for fragmentation and sequencing.
The dependence of charge state on the separation of isomeric species by DMS was also demonstrated. This separation was likely due to the subtle structural differences between the MUC5AC-3 and MUC5AC-13 peptides, which were either highlighted by (or actually induced by) the degree of protonation of these isomers. This interesting observation is presently the subject of further investigation.
References
Apweiler, R., Hermjakob, H., Sharon, N.: On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473, 4–8 (1999)
Harvey, D.J.: Identification of protein-bound carbohydrates by mass spectrometry. Proteomics 1, 311–328 (2001)
Wuhrer, W., Catalina, M.I., Deelder, A.M., Hokke, C.H.: Glycoproteomics based on tandem mass spectrometry of glycopeptides. J. Chromatogr. B 849, 115–128 (2007)
Alving, K., Paulsen, H., Peter-Katalinic, J.: Characterization of O-glycosylation sites in MUC2 glycopeptides by nanoelectrospray QTOF mass spectrometry. J. Mass Spectrom. 34, 395–407 (1999)
Zubarev, R.A., Kelleher, N.L., McLafferty, F.W.: Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 120, 3265–3266 (1998)
Håkansson, K., Cooper, H.J., Emmett, M.R., Costello, C.E., Marshall, A.G., Nilsson, C.L.: Electron capture dissociation and infrared multiphoton dissociation ms/ms of an N-glycosylated tryptic peptide to yield complementary sequence information. Anal. Chem. 73, 4530–4536 (2001)
Syka, J.E., Coon, J.J., Schroeder, M.J., Shabanowitz, J., Hunt, D.F.: Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 (2004)
Mechref, Y.: Use of CID/ETD mass spectrometry to analyze glycopeptides. Curr. Protoc. Protein Sci. 68, 12.11.1–12.11.11 (2012)
Hogan, J.M., Pitteri, S.J., Chrisman, P.A., McLuckey, S.A.: Complementary structural information from a tryptic N-linked glycopeptide via electron transfer ion/ion reactions and collision-induced dissociation. J. Proteome Res. 4, 628–632 (2005)
Han, H., Xia, Y., Yang, M., McLuckey, S.A.: Rapidly alternating transmission mode electron-transfer dissociation and collisional activation for the characterization of polypeptide ions. Anal. Chem. 80, 3492–3497 (2008)
Shvartsburg, A.A.: Differential ion mobility spectrometry: nonlinear ion transport and fundamentals of FAIMS. CRC Press, Boca Raton (2009)
Purves, R.W., Guevremont, R.: Electrospray ionization high-field asymmetric waveform ion mobility spectrometry-mass spectrometry. Anal. Chem. 71, 2346–2357 (1999)
Krylov, E.V., Nazarov, E.G., Miller, R.A.: Differential mobility spectrometer: model of operation. Int. J. Mass Spectrom. 226, 76–85 (2007)
Schneider, B.B., Covey, T.R., Coy, S.L., Krylov, E.V., Nazarov, E.G.: Planar differential mobility spectrometer as a pre-filter for atmospheric pressure ionization mass spectrometry. Int. J. Mass Spectrom. 298, 45–54 (2010)
Barnett, D.A., Ells, B., Guevremont, R., Purves, R.W.: Separation of leucine and isoleucine by electrospray ionization-high field asymmetric waveform ion mobility spectrometry-mass spectrometry. J. Am. Soc. Mass Spectrom. 10, 1279–1284 (1999)
Maccarone, A.T., Duldig, J., Mitchell, T.W., Blanksby, S.J., Duchoslav, E., Campbell, J.L.: Rapid and unambiguous characterization of acyl chains in phosphatidylcholines using differential mobility and mass spectrometry. J. Lipid Res. 55, 1668 (2014)
Hernandez, O., Isenberg, S., Steinmetz, V., Glish, G.L., Maître, P.: Probing mobility-selected saccharide isomers: selective ion–molecule reactions and wavelength-specific IR activation. J. Phys. Chem. A 119, 6057–6064 (2015)
Blagojevic, V., Chramow, A., Schneider, B.B., Covey, T.R., Bohme, D.K.: Differential mobility spectrometry of isomeric protonated dipeptides: modifier and field effects on ion mobility and stability. Anal. Chem. 83, 3470–3476 (2011)
Campbell, J.L., Le Blanc, J.C.Y., Schneider, B.B.L.: Probing electrospray ionization dynamics using differential mobility spectrometry: the curious case of 4-aminobenzoic acid. Anal. Chem. 84, 7857–7864 (2012)
Kovačević, B., Schorr, P., Qi, Y., Volmer, D.A.: Decay mechanisms of protonated 4-quinolone antibiotics after electrospray ionization and ion activation. J. Am. Soc. Mass Spectrom. 25, 1974–1986 (2014)
Campbell, J.L., Yang, A.M.-C., Melo, L.R., Hopkins, W.S.: Studying gas-phase interconversion of tautomers using differential mobility spectrometry. J. Am. Soc. Mass Spectrom. 27, 1277–1284 (2016)
Creese, A.J., Cooper, H.J.: Separation and identification of isomeric glycopeptides by high field asymmetric waveform ion mobility spectrometry. Anal. Chem. 84, 2597–2601 (2012)
Good, D.M., Wirtala, M., McAlister, G.C., Coon, J.J.: Performance characteristics of electron transfer dissociation mass spectrometry. Mol. Cell. Proteom. 6, 1942–1951 (2007)
McLuckey, S.A., Stephenson Jr., J.L.: Ion/ion chemistry of high-mass multiply charged ions. Mass Spectrom. Rev. 17, 369–407 (1998)
Schneider, B.B., Covey, T.R., Coy, S.L., Krylov, E.V., Nazarov, E.G.: Control of chemical effects in the separation process of a differential mobility mass spectrometer system. Eur. J. Mass Spectrom. 16, 57–71 (2010)
Schneider, B.B., Covey, T.R., Coy, S.L., Krylov, E.V., Nazarov, E.G.: Chemical effects in the separation process of a differential mobility/mass spectrometer system. Anal. Chem. 82, 1867–1880 (2010)
Levin, D.S., Miller, R.A., Nazarov, E.G., Vouros, P.: Rapid separation and quantitative analysis of peptides using a new nanoelectrospray differential mobility spectrometer-mass spectrometer system. Anal. Chem. 78, 5443–5452 (2006)
Baba, T., Campbell, J.L., Le Blanc, J.C.Y., Hager, J.W., Thomson, B.T.: Electron capture dissociation in a branched radio-frequency ion trap. Anal. Chem. 87, 785–792 (2015)
Baba, T., Campbell, J.L.: Capturing polyradical protein cations after an electron capture event: evidence for their stable distonic structures in the gas phase. J. Am. Soc. Mass Spectrom. 26, 1695–1701 (2015)
Schneider, B.B., Nazarov, E.G., Londry, F., Vouros, P., Covey, T.R.: Differential mobility spectrometry/mass spectrometry history, theory, design optimization, simulations, and applications. Mass Spectrom. Rev. 35, 687–737 (2016)
Shvartsburg, A.A., Tang, K., Smith, R.D.: High-resolution differential ion mobility separations using helium-rich gases. Anal. Chem. 82, 2456–2462 (2010)
Santiago, B.G., Harris, R.A., Isenberg, S.L., Ridgeway, M.E., Pilo, A.L., Kaplan, D.A., Glish, G.L.: Improved differential ion mobility separations using linked scans of carrier gas composition and compensation field. J. Am. Soc. Mass Spectrom. 26, 1746–1753 (2015)
Seipert, R.R., Dodds, E.D., Clowers, B.H., Beecroft, S.M., German, J.B., Lebrilla, C.B.: Factors that influence fragmentation behavior of N-linked glycopeptide ions. Anal. Chem. 80, 3684–3692 (2008)
Campbell, J.L., Zhu, M., Hopkins, W.S.: Ion-molecule clustering in differential mobility spectrometry: lessons learned from tetraalkylammonium cations and their isomers. J. Am. Soc. Mass Spectrom. 25, 1583–1591 (2014)
Liu, C., Le Blanc, J.C.Y., Shields, J., Janiszewski, J.S., Ieritano, C., Ye, G.F., Hawes, G.F., Hopkins, W.S., Campbell, J.L.: Using differential mobility spectrometry to measure ion solvation: an examination of the roles of solvents and ionic structures in separating quinoline-based drugs. Analyst 140, 6897–6903 (2015)
Liu, C., Le Blanc, J.C.Y., Schneider, B.B., Shields, J., Federico III, J.J., Zhang, H., Stroh, J.G., Kauffman, G.W., Kung, D.W., Ieritano, C., Shepherson, E., Verbuyst, M., Melo, L., Hasan, M., Naser, D., Janiszewski, J.S., Hopkins, W.S., Campbell, J.L.: Assessing physicochemical properties of drug molecules via microsolvation measurements with differential mobility spectrometry. ACS Cent. Sci. 3, 101–109 (2017)
Lintonen, T., Baker, P.R.S., Suoniemi, M., Ubhi, B., Koistinen, K., Duchoslav, E., Campbell, J.L., Ekroos, K.: Differential mobility spectrometry-driven shotgun lipidomics. Anal. Chem. 86, 9662–9669 (2014)
Kjeldsen, F., Haselmann, K.F., Budnik, B.A., Jensen, F., Zubarev, R.A.: Dissociative capture of hot (3–13 eV) electrons by polypeptide polycations: an efficient process accompanied by secondary fragmentation. Chem. Phys. Lett. 356, 201–206 (2002)
Manri, N., Satake, H., Kaneko, A., Hirabayashi, A., Baba, T., Sakamoto, T.: Glycopeptide identification using liquid-chromatography-compatible hot electron capture dissociation in a radio-frequency-quadrupole ion trap. Anal. Chem. 85, 2056–2063 (2013)
Mentinova, M., Crizer, D.M., Baba, T., McGee, W.M., Glish, G.L., McLuckey, S.A.: Cation recombination energy/coulomb repulsion effects in ETD/ECD as revealed by variation of charge per residue at fixed total charge. J. Am. Soc. Mass Spectrom. 24, 1676–1689 (2013)
Acknowledgements
The authors thank Dr. Bradley Schneider (SCIEX) for very helpful discussions and suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Campbell, J.L., Baba, T., Liu, C. et al. Analyzing Glycopeptide Isomers by Combining Differential Mobility Spectrometry with Electron- and Collision-Based Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom. 28, 1374–1381 (2017). https://doi.org/10.1007/s13361-017-1663-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-017-1663-6